Figures & data
Figure 1. (A) Normal pig skin. Viable epidermal cells (star) exhibit minimal autofluorescence that is excluded (black) by the ImageJ filter (C). Dermal collagen (arrow) and superficial keratin are not excluded by the filter and remain white. H&E (A), fluorescent (B), and filtered (C) 400×. (B) Normal pig skin. Viable hair follicle (A,C,E) and mammary gland (B,D,F). Follicular and mammary epithelia (arrows) exhibit minimal autofluorescence that is excluded by the filter. H&E (A,B), fluorescent (C,D) and filtered (E, F) 400× magnification.
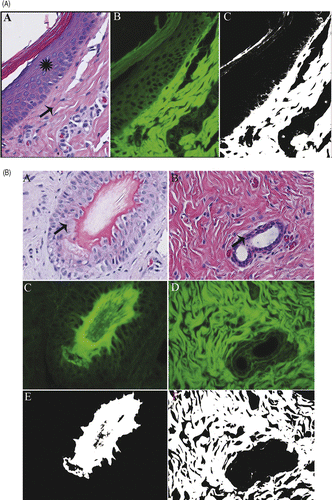
Figure 2. Tumor and skin ablation: Non-viable, ablated skin (A,B,C) and rabbit VX2 carcinoma (D,E,F). Ablated non-viable skin (A,B,C) exhibits increased autofluorescence, and remains white in filtered image. Non-viable tumor cells are strongly autofluorescent, and are white in filtered image. Viable VX2 carcinoma cells (G,H,I) fluoresce less strongly and remain black in filtered image. H&E (A,D,G), fluorescent (B,E,H), filtered (C,F,I), 400× magnification.
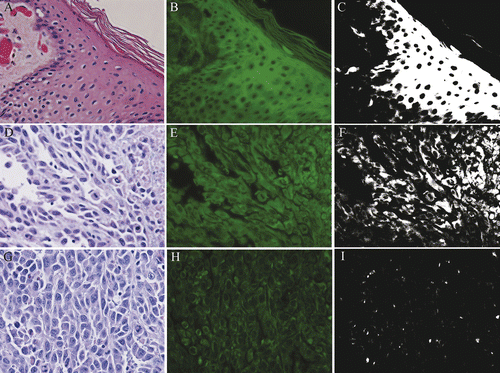
Figure 3. Receiver Operating Characteristics (ROC) analysis of using an autofluorescence intensity (grayscale) threshold to distinguish viable from non-viable samples. The line at top is the empirical ROC curve for classifying Fig 4 data via grayscale threshold; the diagonal is the theoretical ROC curve of a classification rule with no discriminatory potential. The area under the empirical ROC curve is 0.96 (1-sided Mann-Whitney P = 5.6 × 10−5). The ellipse denotes where sensitivity (92%) and specificity (100%) has a sum that is maximized. This optimal combination of 92% sensitivity with 100% specificity is produced by a range of grayscale intensity thresholds that have a minimum of 20.99 and maximum of 25.49.
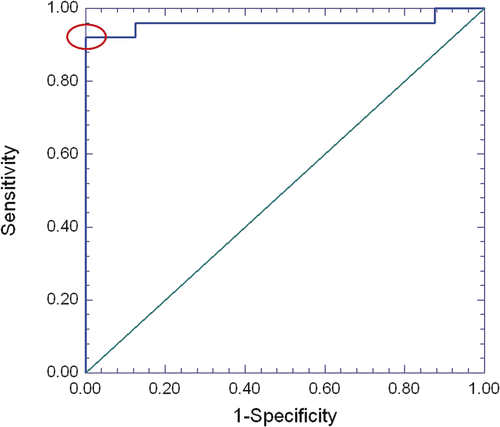
Figure 4. Distribution of autofluorescence grayscale values of viable (n = 5 VX2 v samples and n = 3 non-ablated pig skin samples) and non-viable (heat-killed, n = 25 VX2 carcinoma samples) tissue sections. Open circles denote tissue-section fluorescence intensities, determined as the average grayscale value of single cells in five pre-determined locations in the tissue section; grey diamonds (error bars) denote the sample averages (standard deviations) of fluorescence intensities of the viable and non-viable sections. The vertical line denotes an integer-valued grayscale threshold chosen from the optimal range of thresholds identified by ROC analysis (Fig 3 legend). 23 out of 25 non-viable samples lie above the threshold (92% sensitivity), while all 8 viable samples lie below the threshold (100% specificity).
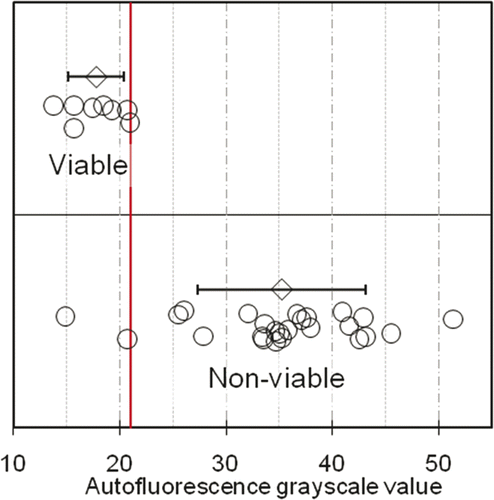
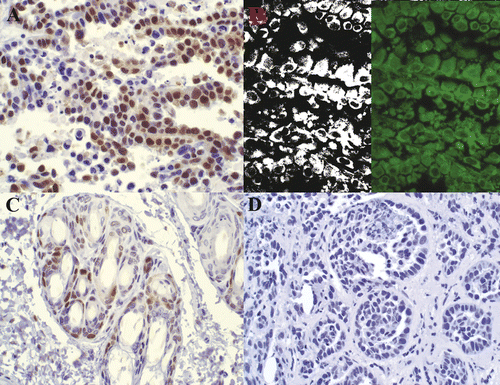
Figure 5. PCNA staining in non-viable tumor cells. Rabbit VX2 carcinoma. Strong nuclear PCNA antibody labeling (A) is observed in tumor cells within a region identified as non-viable via TTC. Fluorescent and filtered images (B) demonstrate strong autofluorescence. Control (C) with primary antibody omitted is unstained. 400× magnification.