Figures & data
Figure 1. Diagnosis of CA. (A) HE staining and pathological examination of the samples dissected from the 3 samples of CA tissue. Arrows indicate partial characteristic pathological features that substantiate the diagnosis. (B) PCR amplification of HPV-DNA samples from the 3 tissue samples were separated by agarose gel electrophoresis. A 100 bp DNA ladder was used to illustrate DNA amplicon length. DNA from HaCaT cells were used as a negative control, whereas DNA from CaSki cells were used as a positive control.
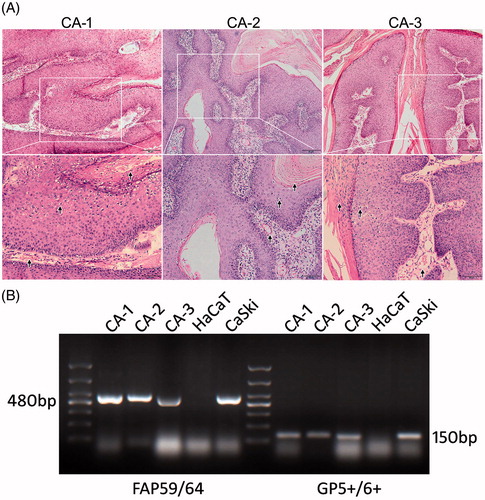
Figure 2. Workflow of the study and Volcano Plot for protein expressions. (A) A brief summary of procedures involved with sample preparation, grouping, iTRAQ labeling, MS detection and bioinformatics analysis used in this study. (B) The Volcano plot for quantifiable proteins identified by LC-MS/MS. Spots on the top-right quadrant indicate significantly upregulated proteins (p values <.05) with a fold change greater than 1.2, spots on the top-left quadrant indicate significantly downregulated proteins (p values <.05) with a fold change less than 0.833 and the other spots indicate proteins failing to meet these criteria of fold change and/or p values.
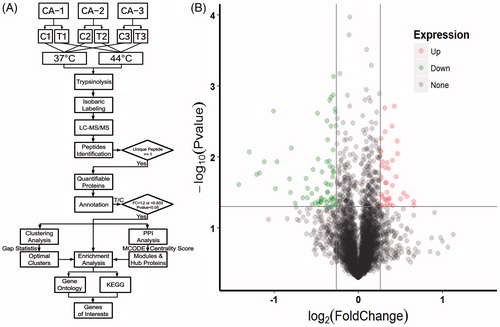
Table 1. The top 10 up- and down-regulated proteins identified from iTRAQ analysis.
Figure 3. Validation of proteomic results on mRNA levels by both real-time qPCR and western blot assay. Relative fold change of mRNA expression levels of OAS1 (A), MX1 (B), AP1S1 (C), BANF1 (D), CANX (E), EXOSC4 (F), GALT (G), KRT5 (H), EXOSC6 (I) and KRT76 (J) are illustrated in the bar plots. Protein expressions of CANX, MX1, GALT, KRT5 and KRT76 were illustrated in (K), and GAPDH was used as a loading control. The grayscale analysis of western blot results was shown in barplot (L). CA samples were assigned to either 37 °C control group (37) or 44 °C hyperthermia (44). Asterisk (*) indicates statistically significant differences between the hyperthermia and control groups, whereas ns indicates non-significant differences.
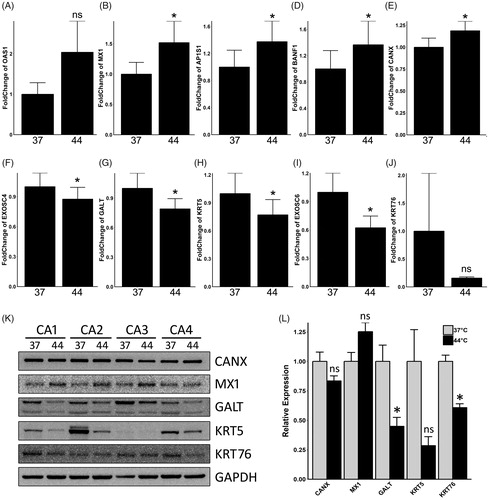
Figure 4. Gene ontology and KEGG enrichment analysis of DEPs. (A) The top ten gene counts of GO-BP (top 10 bars), MF (middle 10 bars) and CC (bottom 10 bars) terms are illustrated on the left side, with respective enrichment scores (−log10 (p values) ×10) shown on the right. (B) The top ten gene counts of KEGG terms are illustrated on the left side, and their respective enrichment scores (−log10 (p values) ×10) are shown on the right.
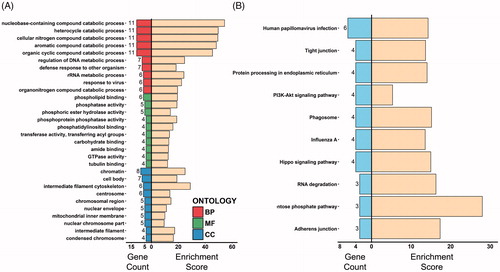
Figure 5. Clustering analysis of the DEPs and GO-BP enrichment of clusters. (A) The optimal cluster number was evaluated with use of gap statistics. Dashed line indicates the optimal k value of 2. (B) K-means clustering analysis of DEPs applying optimal k value revealed 2 distinct clusters, with triangles indicating Cluster 1 and spots Cluster 2. (C) Hierarchical clustering analysis of DEPs applying the optimal k value. ‘C’ Refers to the 37 C control groups whereas ‘T’ refers to the 44 C hyperthermia groups. Gene symbols of DEPs are illustrated on the right side. (D) Biological process terms of the top 15 gene count in the GO-BP enrichment analysis for Cluster 1 (top 15 bars on the left) and Cluster 2 (bottom 15 bars on the left). Bars on the right represent respective enrichment scores (−log10 (p values) ×10).
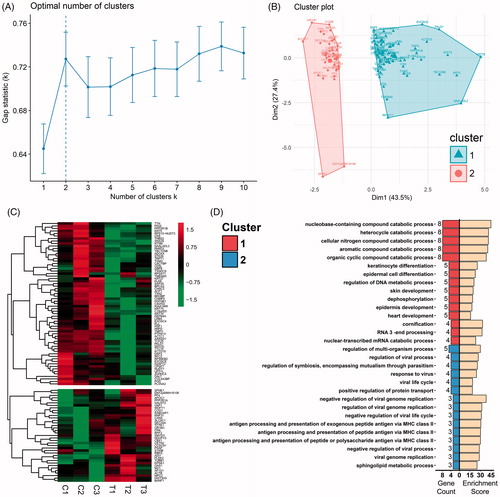
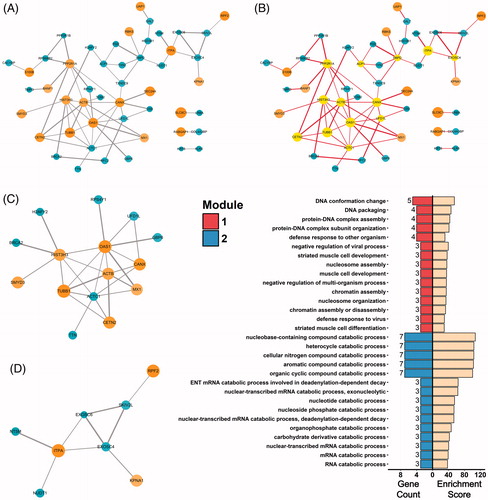
Figure 6. Protein-protein interactions of DEPs. (A) PPI network of DEPs visualized by Cytoscape. Circles indicate up- or down-regulated DEPs. Edges connecting the circles indicate protein-protein interactions, with thickness of the edges representing confidence of the bonds. (B) Hub proteins identified by CentiScaPe were highlighted in the PPI network. (C and D) Interaction module 1 (C) and module 2 (D) mined from the PPI network with use of MCODE analysis. (E) Biological process terms of the top 15 gene count in the GO-BP enrichment analysis for Module 1 (top 15 bars on the left) and Module 2 (bottom 15 bars on the left). Bars on the right represent respective enrichment scores (−log10 (p values) ×10).