Figures & data
Figure 1. Target sequences and chemical structures of the N-methylpyrrole-N-methylimidazole hairpin polyamides. Consensus binding site for HIF is indicated by an open box. Open and closed circles represent imidazole and pyrrole rings, respectively. Dp and b denote dimethylaminopropylamine and b-alanine residues, respectively.
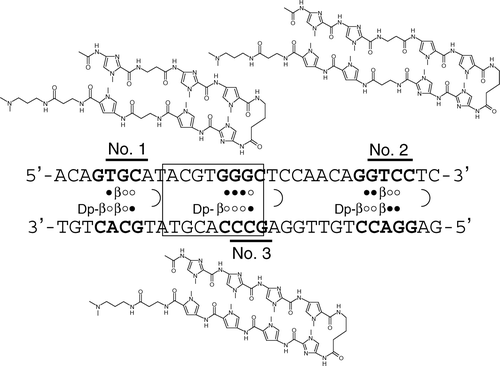
Table I. Molecular weights of polyamides measured by ESIMS.
Figure 2. Dose-dependent inhibition of HIF-1α-induced transcription in 293 cells. Cells were transfected with plasmids coding hypoxia-stable HIF-1α, ARNT, and β-galactosidase, along with a reporter plasmid (pGL3VEGF), and then treated with the mixture of three pyrrole-imidazole hairpin polyamides for 24 h. Transcriptional activity was measured and expressed as a ratio of luciferase to β-galactosidase activity. Statistically significant suppression of transcription was noted at a concentration of 5 µM or greater. Cell viability was not affected by the polyamides. RLU and β-Gal denote relative luciferase units and β-galactosidase activity, respectively. OD: optical density. *: Vehicle (dimethylsulfoxide) only.
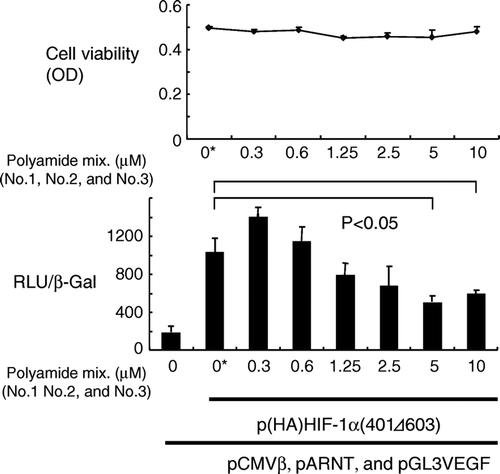
Figure 3. Effects of combinations of the three polyamides on HIF-1α -induced transcription in 293 cells. Cells were transfected with plasmids coding for hypoxia-stable HIF-1α(p(HA)HIF-1α(401Δ603)), ARNT (pARNT), and β-galactosidase (pCMVβ), along with a reporter plasmid (pGL3VEGF), and then incubated with a particular combination of the pyrrole-imidazole polyamides for 24 h. Transcriptional activity was expressed as a ratio of luciferase to β-galactosidase activity. A statistically significant reduction in transcription was achieved by applying all three Py-Im hairpin polyamides. A similar suppression was observed when polyamide no. 3 was applied alone or together with either polyamide no. 1 or no. 2. Cell viability was not affected by the treatment. RLU and β-Gal represent relative luciferase units and β-galactosidase activity, respectively. *: P < 0.05 compared with controls.
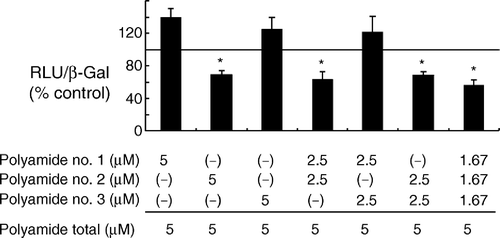
Figure 4. Dose-dependent inhibition of HIF-2α-induced transcription in 293 cells. Cells were transfected with plasmids coding full-length HIF-2α (pTRE-EPAS), ARNT(pARNT), and β-galactosidase (pCMVβ), along with a reporter plasmid (pGL3VEGF), and then treated with a mixture of the three pyrrole-imidazole hairpin polyamides for 24 h. Transcriptional activity was measured and expressed as a ratio of luciferase to β-galactosidase activity. Statistically significant suppression of transcription was noted at a concentration of 2.5 µM or greater. RLU and β-Gal denote relative luciferase units and β-galactosidase activity, respectively. *: Vehicle (dimethylsulfoxide) only.
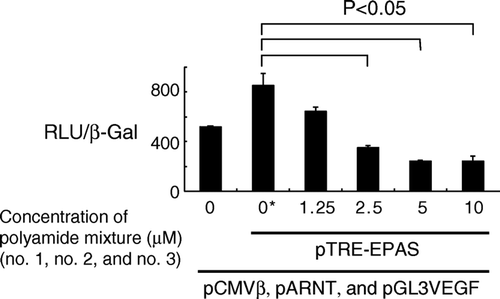
Figure 5. Suppressive effects of the mixture of three Py-Im hairpin polyamides on VEGF transcription and secretion in A498, a renal cell carcinoma cell line. Cells were treated with the mixture of three pyrrole-imidazole polyamides (5 mM in total) for 24–72 h. Transcription of the VEGF gene was evaluated by real time RT-PCR. The results were expressed as a ratio of VEGF mRNA to GAPDH mRNA. An approximately 50% reduction in transcription was observed after 72 h treatment. Consistently, VEGF secreted in conditioned medium (CM) decreased by approximately 50% compared with the control. *: P < 0.05 compared with controls.
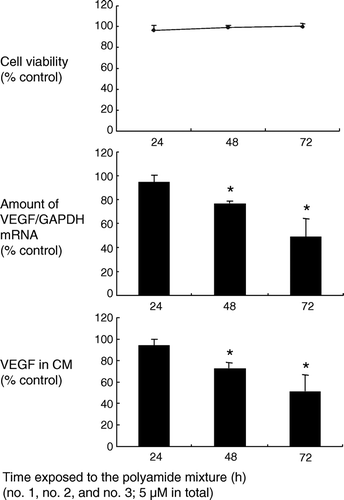
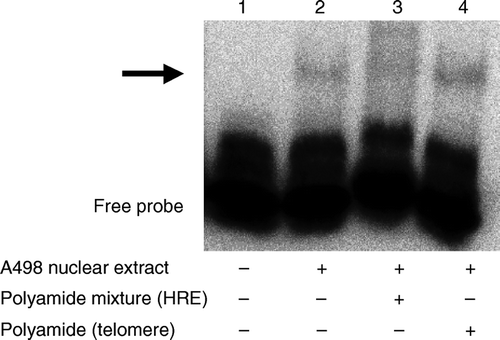
Figure 6. Inhibitory effects of pyrrole-imidazole hairpin polyamides on the HIF-HRE interaction were evaluated by using an electrophoresis mobility shift assay. Nuclear extract of A498 human renal cell cancer cells caused a mobility shift (arrow) of a biotin-labeled double-stranded oligonucleotide corresponding to human HRE (lanes 1 and 2). The polyamide mixture against the HRE suppressed the mobility shift (lane 3), which was not affected by a polyamide recognizing the human telomere sequence (lane 4).