Figures & data
Table 1. Primers used in this study.
Table 2. The names, accession numbers, countries of origin and hosts of SMYEV isolates with full-length coat protein sequences available in the NCBI database.
Fig. 1 Schematic representation of the genomic organization of Strawberry mild yellow edge virus (SMYEV) isolate AB5-2. ORF1 through ORF5 are defined by the vertical number. ORF1 codes for a putative viral replicase, which contains the conserved domains for methyltransferase (MET), viral RNA helicase, and RNA-dependent RNA polymerase (RdRp). The AB5-2 TGBp1 has a conserved helicase domain (nt 4171–4761).

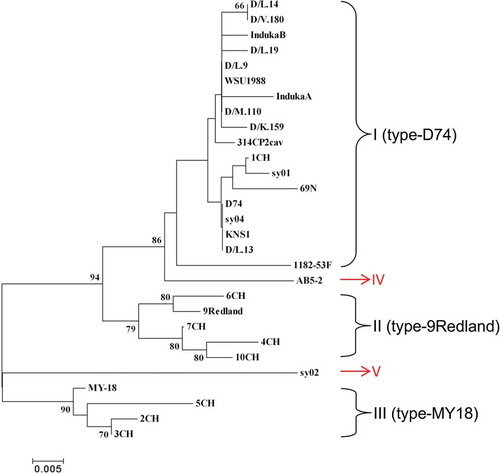
Fig. 2 (Colour online) A phylogenetic tree based on the coat protein (CP) nucleotide sequences of 28 Strawberry mild yellow edge virus (SMYEV) isolates retrieved from the NCBI database and AB5-2 determined in this study. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurring in this group. The scale bar represents the number of substitutions per nucleotide.
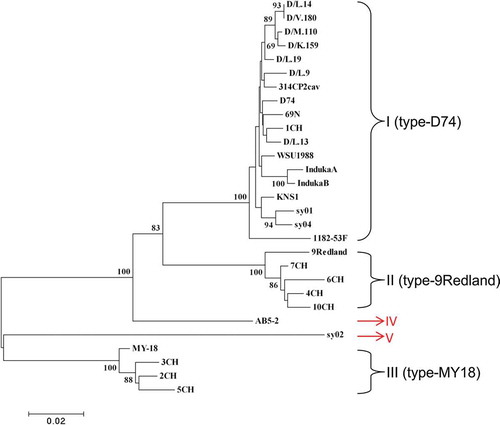
Fig. 3 (Colour online) Phylogenetic tree based on the coat protein (CP) amino acid sequences of 28 Strawberry mild yellow edge virus (SMYEV) isolates retrieved from the NCBI database and the Canadian isolate AB5-2 determined in this study. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurring in this group. The scale bar represents the number of substitutions per amino acid.