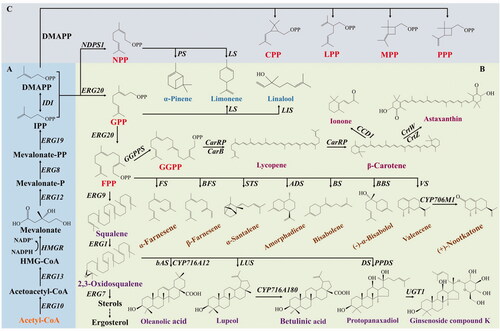
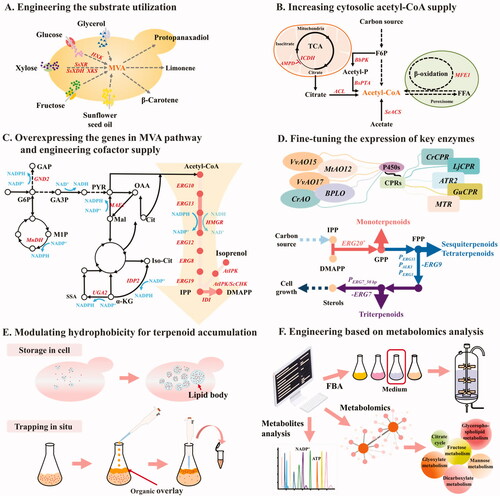
Figures & data
Table 1. Summary of metabolic engineering strategies for improving terpenoids biosynthesis in Y. lipolytica, and information of representative terpenes with the highest titers reported in E. coli and S. cerevisiae.