Figures & data
Figure 1 TheCFH gene contains 20 short consensus repeats (SCR), or complement control modules, each of which encodes a functional domain of ∼60 amino acids. In addition to the full‐length transcript, there is an alternatively spliced, truncated gene product of CFH that has sequence identity with SCRs 1–7. An 8th exon spliced to SCR 7 in the truncated variant (black rectangle) encodes a stop codon, a unique untranslated region, and four C‐terminal amino acids (SFTL). The full length and truncated forms of CFH transcripts are referred to as variants 1 and 2, and the respective encoded proteins are designated as isoforms a and b. The 20 SCRs of CFH encode a number of functional domains. The complement factor I cofactor and decay‐accelerating activities reside within SCRs 1–4 (boxed). Three C3b binding sites are associated with SCRs 1–4, 12–14, and 19–20 (shown in yellow). A RGD sequence in SCR 4 mediates cell adhesion through binding to complement receptor 3 (CR3), an αM integrin. A binding site for C‐reactive protein (CRP), has been localized to the region spanning SCRs 7–11 (blue; in brackets), and a binding site for the Streptococcus pneumoniae surface protein β lies within a domain encompassing SCRs 8–11 (not indicated). Low‐ and high‐affinity adrenomedullin (AM) binding sites have been mapped to SCRs 8–11 and SCRs 15–20 respectively. Three heparin binding sites have been localized to SCRs 7, 12–14, and 20. Also located within SCR 20 is a sialic acid binding site (not indicated). The five related members of the CFH gene family are represented by five separate genes (gene symbols CFHR1–5) all of which reside within the regulators of complement activation (RCA) gene cluster on chromosome 1q32. Each of the 5 CFH‐related genes contains a subset of the 20 CFH SCR domains (see Zipfel, et al., 2002 Citation33 for review). The amino acid sequence homologies of CFHR1–5 SCRs to their CFH counterparts are shown as a percentage in white numerals above each SCR. With the exception of CFHR4 where SCR 7 is absent, all members of the CFH family contain two conserved regions corresponding to SCRs 6–7 and 19–20 of CFH (highlighted in red), and no members contain domains corresponding to SCRs 1–4 of CFH. Based upon their close structural relationships, it may be predicted that the five CFH‐related family members also share functional similarities with CFH.
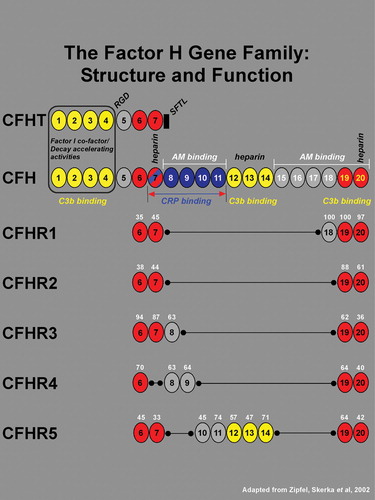
Figure 2 A representative Western blot of serum proteins from 10 of 144 individuals analyzed by nonreducing SDS‐PAGE and chemiluminescent immunoblotting. Upper panel: CFH is visualized as the faster migrating of the two immunoreactive bands (∼150 kDa; identified as CFH by co‐migration with purified CFH; not shown) in the high molecular weight region of the gel/blot. The nature of the higher molecular weight CFH antibody‐reactive band was not determined. The blot depicted was exposed to film for 3 seconds. Lower panel: CFHR1β, a glycosylated isoform of CFHR1, can be distinguished from CFHR1α and CFHT/FHL‐1 which comigrate following separation of proteins in the lower molecular weight region of the gel/blot. No immunoreactive CFHR1β is present in the serum of three patients (lanes 2, 6 and 10), two unaffected patients possessing 1277TT genotypes (protective CFH H4 haplotype) and one patient with a 1277CT genotype, respectively. Subsequent SSCP analysis and direct DNA sequencing showed that all three individuals have both a homozygous deletion of the CFHR1 and CFHR3 genes. The blot was exposed to film for 7 minutes.
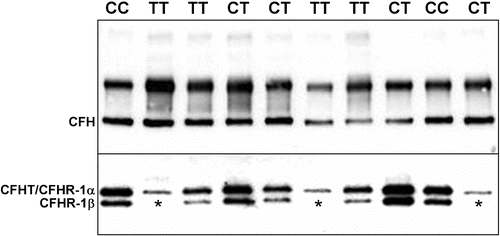
Table I. FHR1 deletion frequencies (cases and controls).
Table II. Ocular CFH, CFHT, CFHR1 and CFHR3 gene expression.
Figure 3 A haplotype network resulting from the analysis of variants in theCFH locus is shown. The size of the sphere is proportional to the estimated frequency of the haplotype. Haplotypes adjacent to each other are predicted to arise from a single mutational step. The small spheres represent nodes that were not observed, but are predicted. The 402H haplotype is depicted, along with the two major protective haplotypes, and the haplotypes that are neutral for AMD risk.
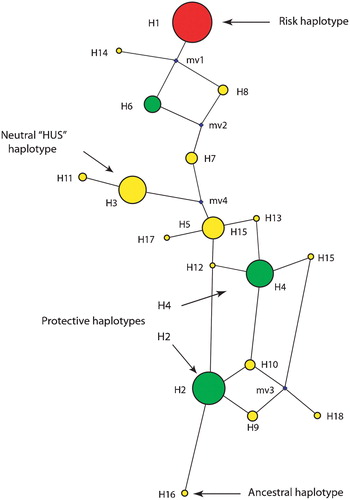
Table III. Population frequencies of CFH T1277C and delCFHR1/CFHR3.
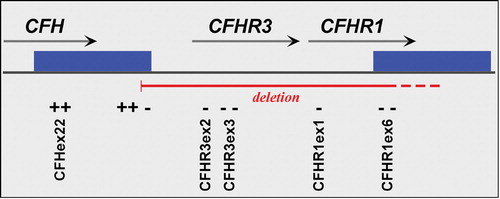
Figure 4 A schematic map of that portion of the RCA cluster on 1q31 showing the relative positions of theCFH, CFHR1 and CFHR3 genes. The solid green boxes represent a segmental duplication of 28 kb that occurred within this locus. The primers employed to define the CFHR1/CFHR3 deletion boundaries are shown in Supplemental Table I; ‘+’ indicates products that amplify in deletion homozygotes, and ‘–’ regions that fail to amplify in deletion homozygotes.