Figures & data
Table 1. Subject characteristics-discovery cohort.
Table 2. Primer sequence for qRTPCR experiment.
Figure 1. Differentially expressed circRNA, miRNA and mRNA in normal and SLE samples. A, differentially expressed cirRNA in GSE108340. B, differentially expressed miRNA in GSE37426. C, differentially expressed mRNA in GSE153781.
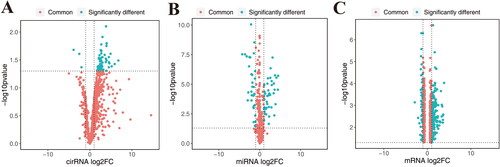
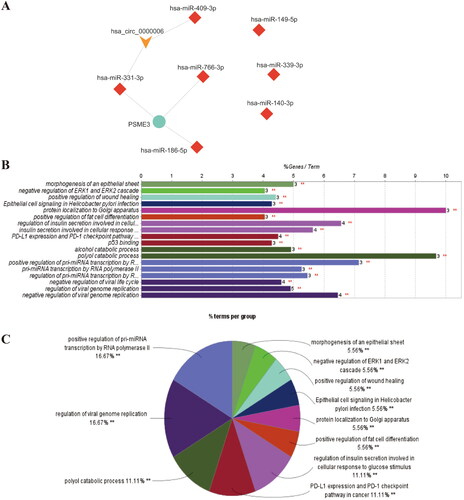
Figure 2. Prediction, construction and analyzation of miRNA-mRNA network. (A) Intersection of target mRNA predicted by miRDB, miR-TarBase and TargetScan databases (B) The cirRNA-miRNA-mRNA ceRNA network; (C) The top 10 hub genes of the ceRNA network.
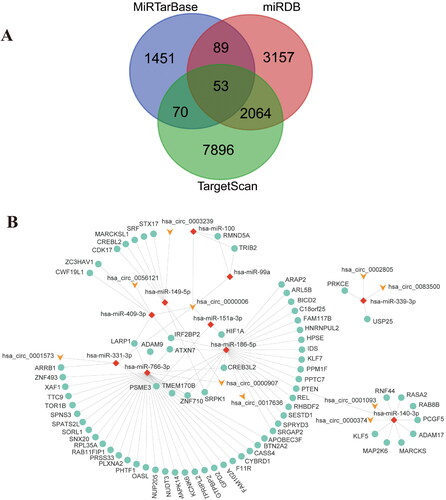
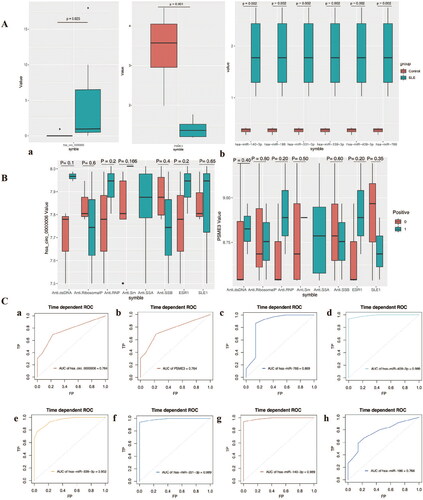
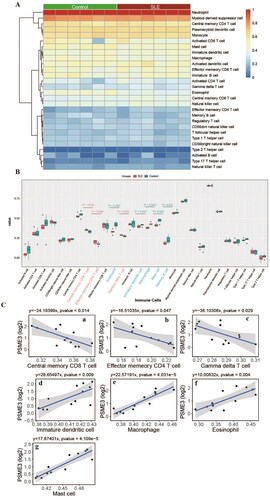
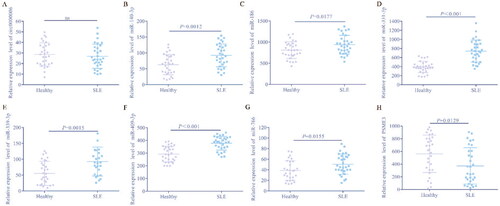
Figure 4. Clinical significance and diagnostic efficiency of hub gene expression. A, Comparison of hub genes expression between SLE patients and control. B, Correlation between the expression of Hub gene and clinicopathological parameters of SLE. a, Correlation between hsa-circ-0000006 (hsa-circRNA-001621) and clinicopathological parameters of SLE; b, Correlation between PSME3 and clinicopathological parameters of SLE. C, Diagnostic efficiency of Hub gene for SLE. a, has_circ_0000006. b, PSEME3. c, hsa-miR-766 = 0.869. d, hsa-miR-409-3p. e, hsa-miR-339-3p. f, hsa-miR-331-3p. g, hsa-miR-140-3p. h, hsa-miR-186-5p.