Figures & data
Figure 1. Pol ζ is highly expressed in glioma and correlated to poor prognosis. The mRNA expression of (A) REV3L and (B) REV7 in 40 paired glioma tissues and paracancerous tissues. n = 40. (C) The protein levels of REV3L and REV7 in tumor and adjacent normal tissues. n = 3. The mRNA expression of (D) REV3L and (E) REV7 in GBM cells (A172, LN229, and U251) and normal cells (NHAs). n = 3. (F) The protein levels of REV3L and REV7 in A172, LN229, U251, and NHAs. n = 3. The prognosis of patients with glioma has high or low (G) REV3L and (H) REV7 expression. n = 40. The expression of REV3L and Rev7 in tissues was assessed using student’s t-test. The prognosis of patients with glioma was evaluated using the Kaplan-Meier analysis. Other comparisons were analyzed using one-way ANOVA. ***p < 0.001.
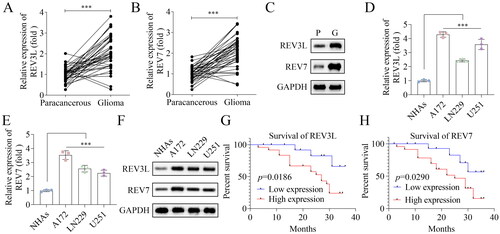
Figure 2. Effects of pol ζ on the radiosensitivity of GBM cells. (A) The efficiency following transfection with (A) si-REV3L 1#, si-REV3L 2#, and si-NC. (B) The efficiency following transfection with si-REV7 1#, si-REV7 2#, and si-NC. (C) Cell viability of transfected cells treated with γ-ray. (D,E) Cell survival was measured after A172 cell transfection and irradiation. (F,G) Cell apoptosis of transfected cells after irradiation. n = 3 in all experiments. The comparisons were analyzed using one-way ANOVA. ***p < 0.001.
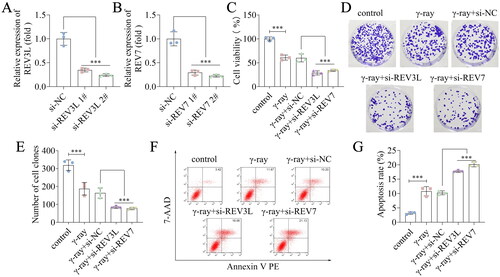
Figure 3. Effects of pol ζ on the immune escape of GBM cells. (A,B) The apoptosis of CD8+ T cells after transfection and irradiation. (C) The protein levels of PD-L1 in A172 cells. n = 3 in all experiments. The comparisons were analyzed using one-way ANOVA. ***p < 0.001. *p < 0.05.
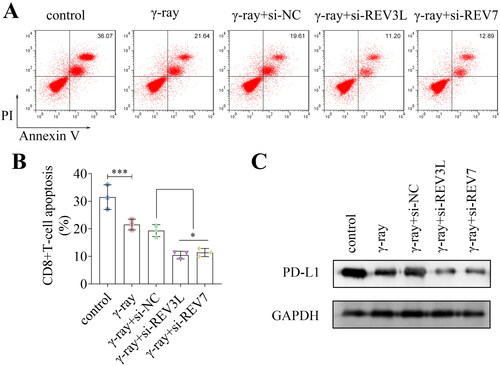
Figure 4. Effects of pol ζ on the PI3K/AKT/mTOR pathway. (A) The protein levels of PI3K, p-PI3K, AKT, p-AKT, mTOR, and p-mTOR in A172 cells treated with γ-ray and transfected with si-REV3L and si-REV7. The ratios of (B) p-PI3K/PI3K, (C) p-AKT/AKT, and (D) p-mTOR/mTOR were quantified. n = 3 in all experiments. The comparisons were analyzed using one-way ANOVA. **p < 0.01.
Figure 5. REV3L affects the radiosensitivity via the PI3K/AKT/mTOR pathway. (A) Cell viability of si-REV3L transfected cells treated with IGF-1. (B,C) Cell survival was measured after transfection and IGF-1 treatment. (D,E) Cell apoptosis of transfected cells after IGF-1 treatment. (F) the protein levels of PI3K, p-PI3K, AKT, p-AKT, mTOR, and p-mTOR. n = 3 in all experiments. The comparisons were analyzed using one-way ANOVA. ***p < 0.001. **p < 0.01.
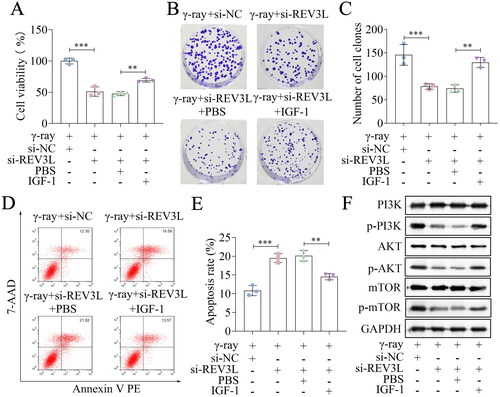
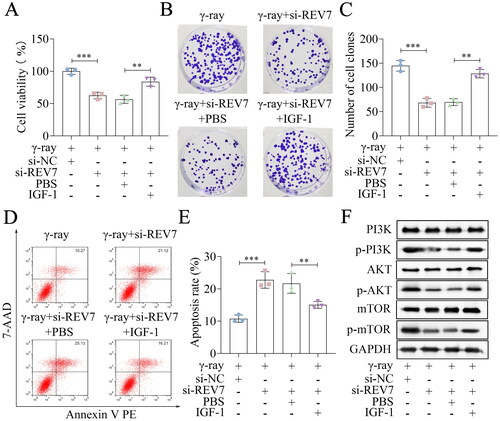
Figure 6. REV7 affects the radiosensitivity via the PI3K/AKT/mTOR pathway. (A) Cell viability of si-REV7 transfected cells treated with IGF-1. (B,C) Cell survival was measured after transfection and IGF-1 treatment. (D,E) Cell apoptosis of transfected cells after IGF-1 treatment. (F) the protein levels of PI3K, p-PI3K, AKT, p-AKT, mTOR, and p-mTOR. n = 3 in all experiments. The comparisons were analyzed using one-way ANOVA. ***p < 0.001. **p < 0.01.
Supplemental Material
Download MS Word (679.5 KB)Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
