Figures & data
Table 1. Primer sequence.
Figure 1. CLDN1 is highly expressed in patients with asthma and PDGF-BB-treated ASMCs. (A) Differentially expressed genes in ASM of patients with fatal asthma and control donors were predicted using microarray. CLDN1 expression in (B) serum samples of patients with asthma and (C) ASMCs treated with PDGF-BB evaluated by RT-qPCR. (D) Protein levels of CLDN1 in ASMCs measured by western blot. All experiments were repeated three times except for the detection of CLDN1 expression in serum samples (n = 28). Data are shown as mean ± standard deviation. **p < 0.01.
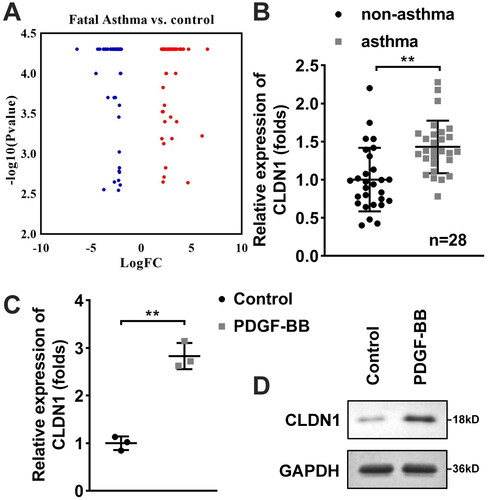
Figure 2. CLDN1 modulates the cellular functions of ASMCs. (A) CLDN1 expression determined by RT-qPCR in ASMCs after transfection. (B) Cell viability of ASMCs evaluated by CCK-8 assay. (C) IL-4, (D) IL-13, and (E) IL-5 levels in ASMCs regulated by CLDN1. (F) The images of ASMCs of EdU and transwell assays. Quantitative analysis of (G) EdU and (H and I) transwell assays. All experiments were repeated three times. Data are shown as mean ± standard deviation. *p < 0.05, **p < 0.01.
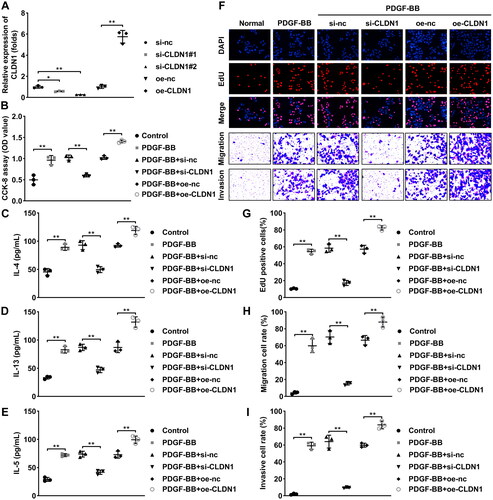
Figure 3. MMP14 is a downstream gene of CLDN1. (A) CLDN1-related proteins predicted by the STRING database. (B) KEGG pathway analysis of the CLDN1-related proteins. (C) mRNA expression of MMP14 after CLDN1 overexpression and knockdown. (D) Fluorescence in situ hybridization assay was performed to analyze the location of CLDN1 and MMP14. (E) Co-IP was carried out to verify that MMP14 could interact with CLDN1. (F) mRNA expression of MMP14 in serum samples of asthma patients. (G) Soluble MMP14 levels in the serum of patients with asthma. (I) Pearson analysis of CLDN1 expression and MMP14 levels in serum of patients with asthma. (H) mRNA and (J) protein levels of MMP14 in ASMCs treated by PDGF-BB. All experiments were repeated three times except for the detection of MMP14 mRNA expression and soluble MMP14 levels in serum samples (n = 28). Data are shown as mean ± standard deviation. **p < 0.01.
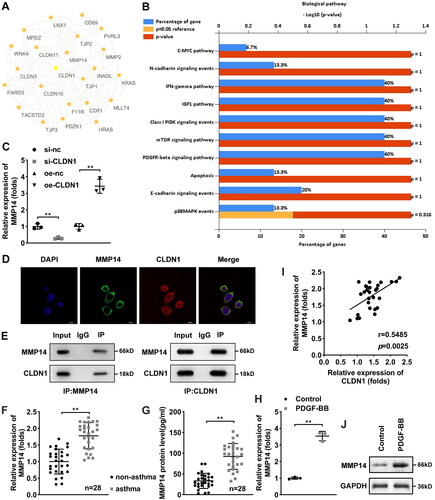
Figure 4. CLDN1 upregulates MMP14 to modulate ASMC growth. (A) MMP14 was successfully elevated in ASMCs. (B) Cell viability of ASMCs evaluated by CCK-8 assay. (C) IL-4, (D) IL-13, and (E) IL-5 levels in ASMCs when CLDN1 knockdown and MMP14 overexpression. (F) The images of ASMCs of EdU and transwell assays. Quantitative analysis of (G) EdU and (H and I) transwell assays. All experiments were repeated three times. Data are shown as mean ± standard deviation. *p < 0.05, **p < 0.01.
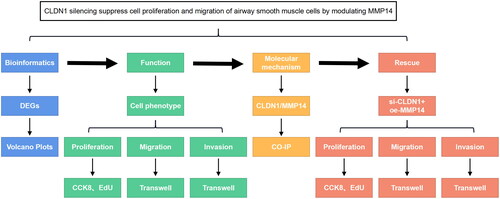
Figure 5. The experimental line of this study. CLDN1 was predicted and confirmed to be dysregulated in asthma. The functions of ASMCs, including proliferation, migration, and invasion, were evaluated. Then, the molecular mechanisms of CLDN1 were identified, which were confirmed using rescue experiments.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
