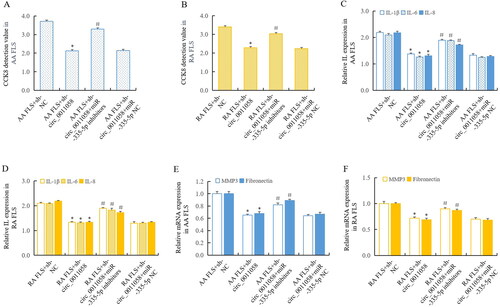
Figures & data
Figure 1. Circ_0011058 expression significantly increased in AA rats and RA FLS.
(A and B) RT-qPCR showed that circ_0011058 was upregulated in synovium and FLS of AA rats. (C) Circ_0011058 was significantly upregulated in RA FLS, further confirming the expression change of circ_0011058 in RA. *AA/RA group vs normal, n = 3 (3 different samples). AA, adjuvant arthritis; RA, rheumatoid arthritis; RT-qPCR, real time qPCR; FLS, fibroblast-like synoviocytes
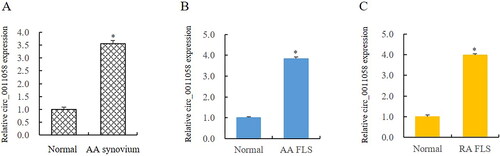
Figure 2. Circ_0011058 knockdown inhibited RA pathology.
CCK8 detection showed that sh-circ0011058 inhibited the proliferation of AA FLS (A) and RA FLS (B). ELISA showed that sh-circ0011058 inhibited the expression of IL-1β, IL-6, and IL-8 in AA FLS (C) and RA FLS (D). RT-qPCR showed that sh-circ0011058 inhibited the expression of MMP3 and fibronectin in AA FLS (E), and also inhibited these two RA genes in RA FLS (F). *AA FLS/RA FLS vs normal, # group 3 mean vs group 2 mean (from left to right), n = 3.
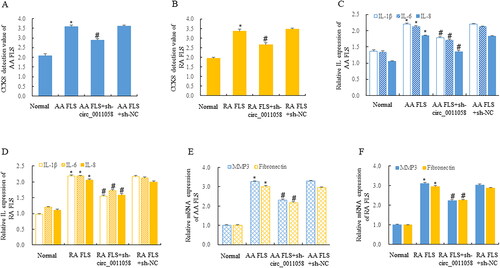
Figure 3. Circ_0011058 directly targeted miR-335-5p.
(A, B) Bioinformatics predicted that miR-335-5p was the direct target of circ_0011058.
(C, D) The expression of miR-149-5p and miR-183-5p was not significantly affected, and the expression of miR-335-5p was significantly upregulated. (E, F) Circ_0011058 knockdown upregulated miR-335-5p expression. Double luciferase reporter gene detection showed that miR-335-5p was the direct target of circ_0011058 (G and H), and RIP experiment confirmed this discovery (I, J). *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3. RIP, RNA Immunoprecipitation
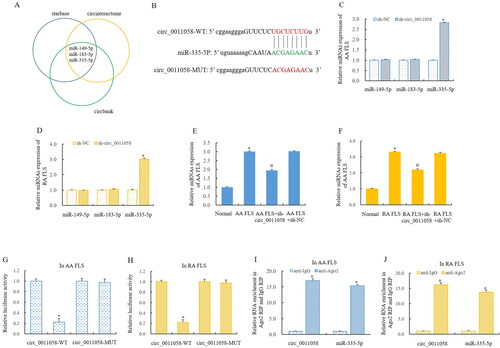
Figure 4. CUL4B was a target of miR-335-5p.
(A) Bioinformatics predicted that CUL4B may be the target of miR-335-5p. RT-qPCR showed that CUL4B expression was significantly upregulated in AA rats (B and C) and RA FLS (D). (E) MiR-335-5p mimics significantly inhibited CUL4B expression. Double luciferase reporter gene detection showed that miR-335-5p directly regulated the CUL4B in AA FLS (F) and RA FLS (G), and RIP detection confirmed the findings (H). *The mean of group 2 vs the mean of group 1, n = 3.
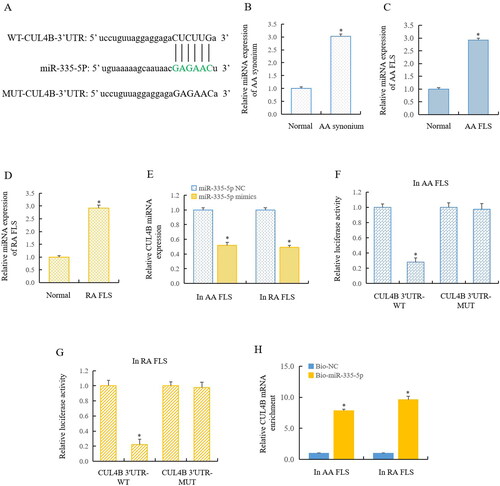
Figure 5. MiR-335-5p mediated the effect of circ_0011058 on CUL4B and canonical Wnt signalling pathway.
RT-qPCR showed that sh-circ_0011058 inhibited CUL4B expression in AA FLS (A) and RA FLS (B), and miR-335-5p inhibitors reversed these effects. WB showed that sh-circ_0011058 inhibited the expression of β-catenin in the nuclei of AA FLS (C) and RA FLS (D), and miR-335-5p inhibitors reversed. (C-1) Quantitative results of β-catenin in AA FLS. (D-1) Quantitative results of β-catenin in RA FLS. (E and F) Sh-circ_0011058 inhibited c-Myc expression, and miR-335-5p inhibitors could reverse them. (G and H) Sh-circ_0011058 inhibited CCND1 expression and miR-335-5p inhibitors reversed. *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3.
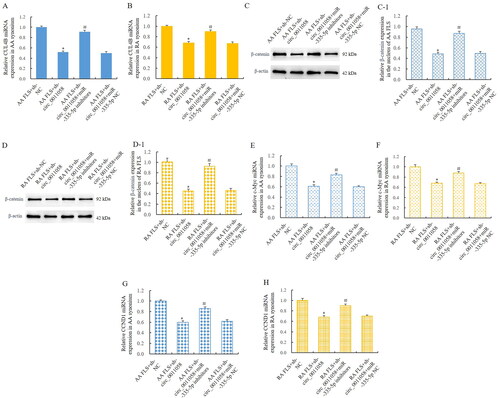
Figure 6. MiR-335-5p mediated the influence of circ_0011058 on RA pathology.
CCK8 showed that sh-circ_0011058 inhibited FLS proliferation in AA FLS (A) and RA FLS (B), and miR-335-5p inhibitors reversed. ELISA showed that sh-circ_0011058 inhibited the expression of IL-1β, IL-6, and IL-8 in AA FLS (C) and RA FLS (D), and miR-335-5p inhibitors reversed. E. F, sh-circ_0011058 inhibited the expression of MMP3 and fibronectin, and miR-335-5p inhibitors could reverse them. *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3.