Figures & data
Table 1. The clinical and demographic characteristics of the patients with TAD and NA.
Table 2. Primer sequences used for qRT-PCR.
Figure 1. CircNRIP1 expression in TAD patients and Ang II-induced HA-VSMCs. (A) Volcano plots showed the differentially expressed circRNAs in TAD patients and NA in GSE97745 dataset. (B) The basic information of circNRIP1. (C) After amplified by the cDNA or gDNA using convergent primers or divergent primers, PCR products were used for agarose gel electrophoresis. (D) RNase R treatment was used to detect the resistance of circNRIP1 on RNase R digestion. (E) Subcellular localisation assay was used to analyse circNRIP1 expression in cytoplasm and nucleus. (F) CircNRIP1 expression in the aortic specimens of TAD patients and NA was analysed by qRT-PCR. (G) CircNRIP1 expression in HA-VSMCs treated with or without Ang II was detected by qRT-PCR. *p < 0.05, **p < 0.01, ***p < 0.001.
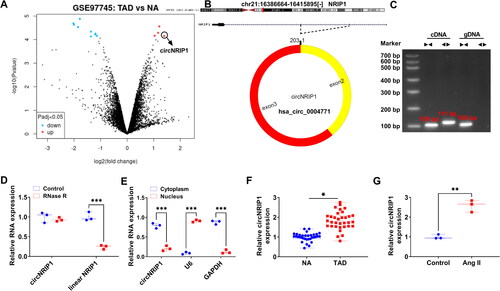
Table 3. Correlation of circNRIP1 expression with clinicopathologic features in patients of TAD.
Figure 2. Effect of si-circNRIP1 knockdown on Ang II-induced HA-VSMCs functions. (A) The transfection efficiencies of si-circNRIP1#1 and si-circNRIP1#2 were detected by qRT-PCR. (B-E) HA-VSMCs were transfected with si-NC/si-circNRIP1 followed by induced with Ang II. EdU assay (B), transwell assay (C) and wound healing assay (D) were used to measure cell proliferation and migration. (E) Protein levels of MMP9, CX43, α-SMA and SM22α were assessed by WB analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
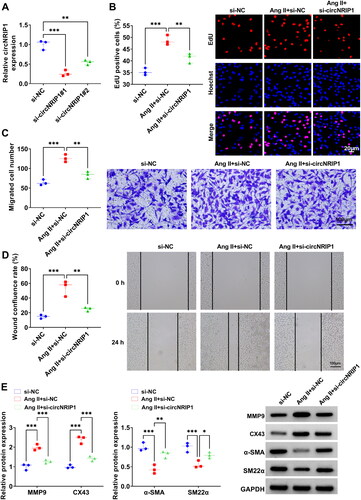
Figure 3. CXCL5 expression in TAD patients and Ang II-induced HA-VSMCs. (A) Volcano plots showed the differentially expressed mRNAs in TAD patients and NA in GSE107844 dataset. (B) CXCL5 mRNA expression in the aortic specimens of TAD patients and NA was detected by qRT-PCR. (C) Pearson correlation coefficient was used for assessing the correlation of CXCL5 mRNA expression and circNRIP1 expression in TAD patients. (D) CXCL5 protein expression was detected by WB analysis in the aortic specimens of TAD patients and NA. (E-F) CXCL5 mRNA and protein expression in HA-VSMCs treated with or without Ang II was examined by qRT-PCR and WB analysis. (G-H) CXCL5 mRNA and protein expression was tested by qRT-PCR and WB analysis in Ang II-induced HA-VSMCs transfected with si-NC/si-circNRIP1. *p < 0.05, **p < 0.01, ***p < 0.001.
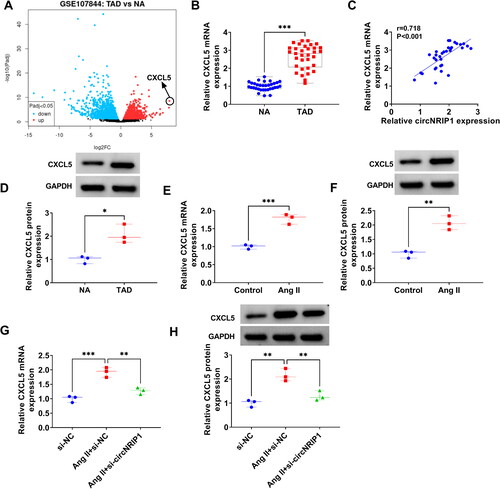
Figure 4. Effect of si-CXCL5 on Ang II-induced HA-VSMCs functions. (A) The transfection efficiencies of si-CXCL5#1 and si-CXCL5#2 were assessed by WB analysis. (B-E) HA-VSMCs were transfected with si-NC/si-CXCL5 followed by induced with Ang II. Cell proliferation and migration were determined by EdU assay (B), transwell assay (C) and wound healing assay (D). (E) WB analysis was used to test the protein levels of MMP9, CX43, α-SMA and SM22α. *p < 0.05, **p < 0.01, ***p < 0.001.
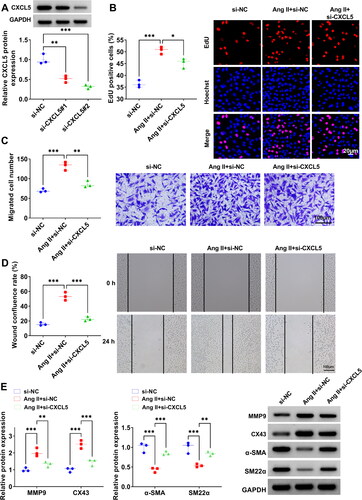
Figure 5. Effects of si-circNRIP1 and oe-CXCL5 on Ang II-induced HA-VSMCs functions. (A) The transfection efficiency of oe-CXCL5 was evaluated by WB analysis. (B-F) HA-VSMCs were transfected with si-NC/si-circNRIP1/vector/oe-CXCL5 followed by induced with Ang II. (B) CXCL5 protein expression was examined by WB analysis. EdU assay (C), transwell assay (D) and wound healing assay (E) were performed to detect cell proliferation and migration. (F) Protein levels of MMP9, CX43, α-SMA and SM22α were measured using WB analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
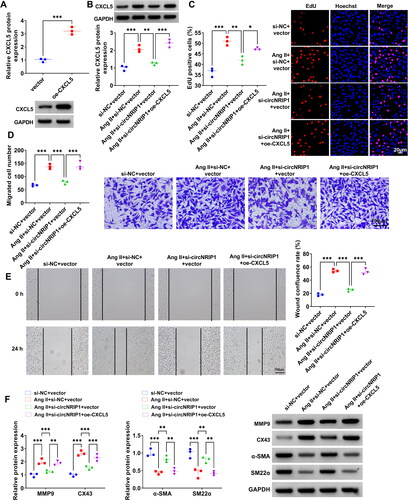
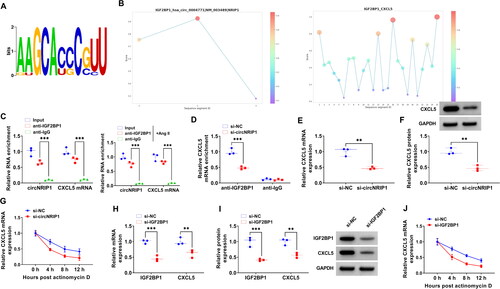
Figure 6. Effect of circNRIP1 and IGF2BP1 on CXCL5 mRNA stability. (A) The known motif of IGF2BP1. (B) RBPsuite predicted the binding sites between IGF2BP1 and circNRIP1/CXCL5. (C-D) RIP assay was used to confirm the interaction between IGF2BP1 and circNRIP1/CXCL5. (E-F) CXCL5 mRNA and protein expression in HA-VSMCs transfected with si-NC/si-circNRIP1 was tested by qRT-PCR and WB analysis. (G) Actinomycin D assay was used to detect the mRNA stability of CXCL5 in HA-VSMCs transfected with si-NC/si-circNRIP1. (H-I) The mRNA and protein levels of IGF2BP1 and CXCL5 were measured by qRT-PCR and WB analysis in HA-VSMCs transfected with si-NC/si-IGF2BP1. (J) The mRNA stability of CXCL5 in HA-VSMCs transfected with si-NC/si-IGF2BP1 was assessed by Actinomycin D assay. **p < 0.01, ***p < 0.001.