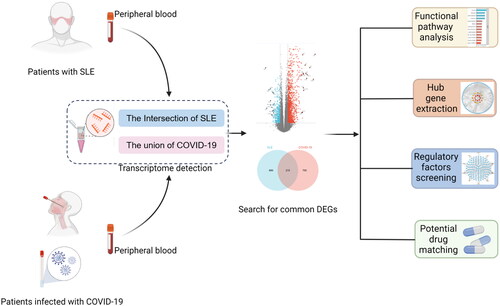
Figures & data
Figure 2. Visualisation the number of common differentially expressed genes between two datasets, COVID-19 and SLE. (A) Comparing the number of differentially expressed genes between COVID-19 and SLE. (B) The volcano plot of differentially expressed genes in 5 datasets. (C) Venn plot of the results of cross-analysis of the three COVID-19 data sets. (D) Venn plot of the cross-analysis results of the 2 SLE datasets. (E) Venn diagram showing the overlap of differentially expressed genes between COVID-19 and SLE.
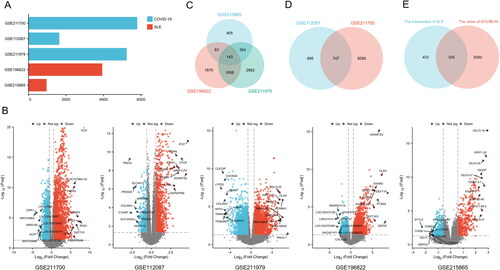
Table 1. Matching of drug candidates.
Figure 3. GO and KEGG functional enrichment analysis of the DEGs between COVID-19 and SLE. (A) The bubble graphs of GO enrichment analysis of common DEGs. (B) The bar graphs of KEGG enrichment analysis of common DEGs.
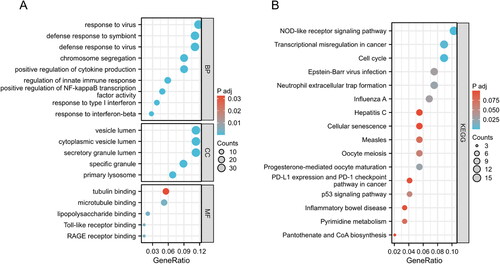
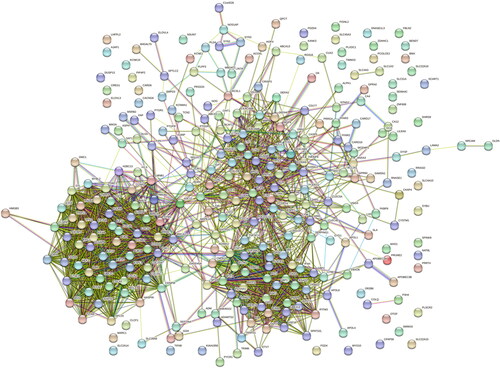
Figure 5. Identification of the top 20 hub genes ranked by their scores. In the middle circle, the 20 central genes represent the top 20 hub genes ranked by their scores and their interactions with other common DEGs.
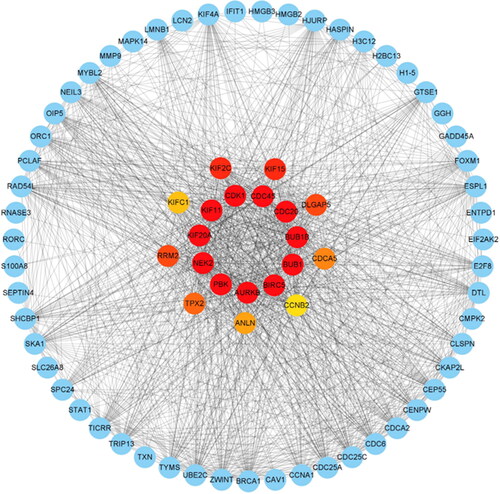
Figure 6. The top 10 transcription factors and their interactions with common DEGs. In this network, the pink squares represent transcription factors with the top 10 ranking, while the blue circles represent common DEGs correlated with transcription factors.
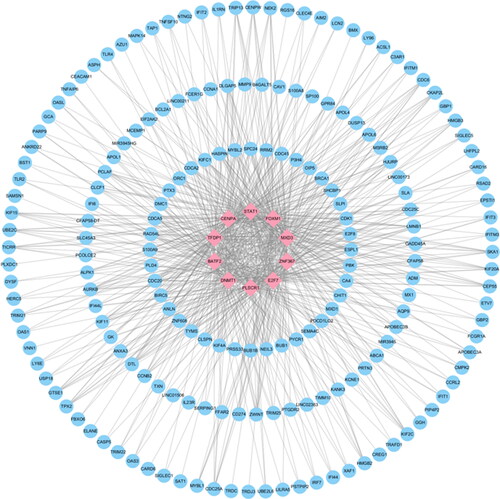
Figure 7. The top 10 miRNAs ranked by their scores and their interactions with common DEGs. In this network, the yellow squares represent miRNAs with the top 10 highest score, while the blue circles represent common DEGs correlated with miRNAs.
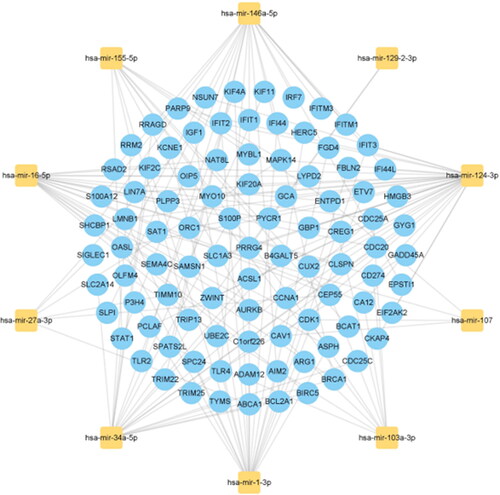
Figure 8. Identification of the top 30 hub genes of DEGs of COVID-19 ranked by their scores. In the middle circle, the central genes represent the top 30 hub genes ranked by their scores and their interactions with other DEGs of COVID-19.
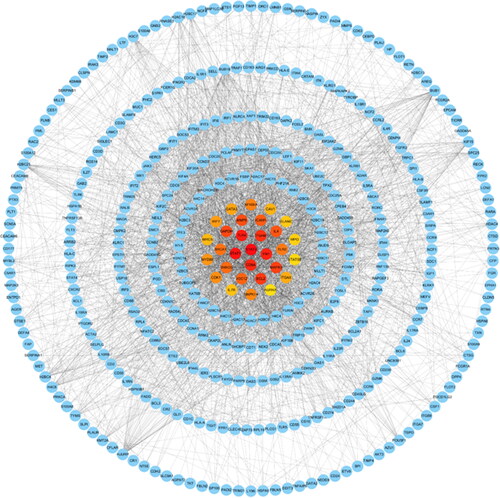
Table 2. Target genes of SLE drugs in COVID-19 and SLE.