Figures & data
Fig. 1. Suppression of FUL1 and FUL2 Inhibited Tomato Fruit Ripening.
Note: (A) FUL1/FUL2 suppressed tomatoes at the age corresponding to the red coloring stage of AC fruits. Scale bars indicate 10 mm. (B) Expression of FUL1, FUL2, and RIN in the suppressed tomatoes. Three suppression lines (TF2, TF18, and TF68) were analyzed, along with a rin mutant and the wild-type (Ailsa Craig, AC), by quantitative RT-PCR (qRT-PCR). Levels of transcripts are shown as fold-change values as compared to the expression in AC at 45 DAP. Error bars indicate standard deviations (SD) from the experiments for three independent fruits.
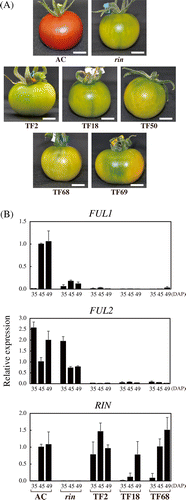
Fig. 2. Inhibition of Ethylene Synthesis by Suppression of FUL1/FUL2.
Note: (A) Ethylene production of FUL1/FUL2 suppressed tomatoes. Fruits of AC, the rin mutant, and three transgenic lines (TF2, TF18, and TF68) were harvested at 40 DAP, and ethylene levels were measured everyday. (B) Expression of ACS2 and ACS4 in the suppressed tomatoes. Expression analyses by qRT-PCR were conducted as described in Fig. .
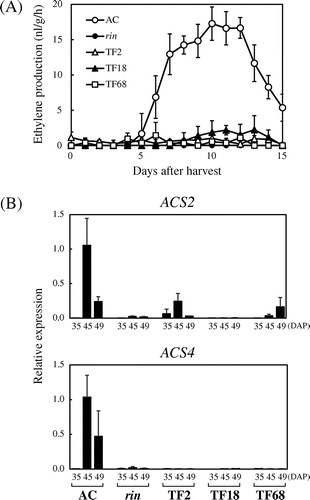
Fig. 3. Inhibition of Lycopene Production by Suppression of FUL1/FUL2.
Note: (A) Lycopene contents in the transgenic tomatoes. Lycopene was measured for the same fruits used to measure ethylene production (15 d after harvest). ND, not detected. (B) Expression of PSY1 in the suppressed tomatoes. Expression analyses by qRT-PCR were conducted as described in Fig. .
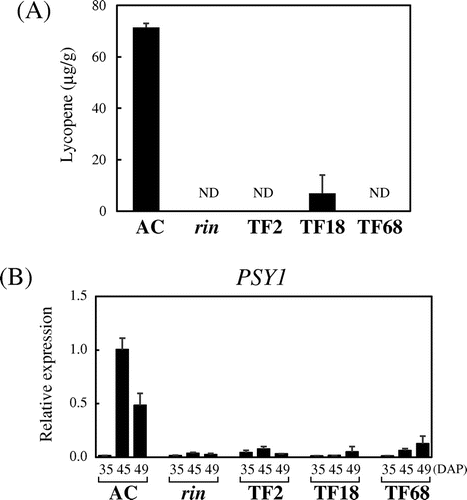
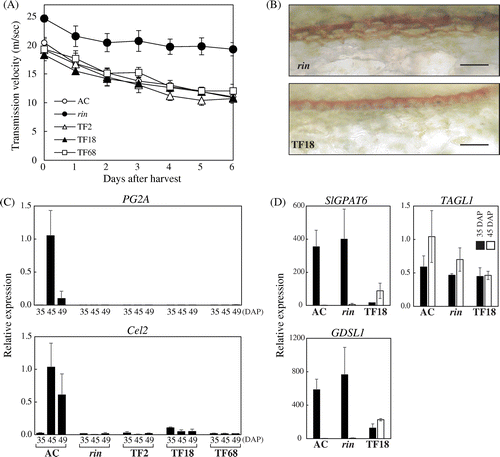
Fig. 4. Reduced Fruit Firmness by FUL1/FUL2 Suppression.
Note: (A) Measurement of fruit firmness of the FUL1/FUL2 suppression lines. Breaker stage fruits, which started coloring (red for AC and yellow for the transgenic lines and the rin mutants), were harvested and firmness was measured everyday (see “Materials and methods”). (B) Expression of PG2A and Cel2 in the fruits of the suppression lines. Expression analyses by qRT-PCR were conducted as described in Fig. . Error bars indicate SD for experiments on three independent fruits in all assays, except for the assay of Cel2 expression in AC at 45-DAP, which used duplicate samples. (C) Cuticle in the peel of the rin mutant and the TF18 transgenic fruits. Cuticular lipids in the fruit tissues were stained with Sudan Red 7B. Scale bars indicate 50 μm. (D) Expression of SlGPAT6, GDSL1, and TAGL1 in the fruits of the FUL1/FUL2 suppression lines. Expression in TF18 at 35 DAP and 45 DAP was analyzed in the rin mutant and the wild-type by qRT-PCR. Levels of transcripts are shown as fold-change values as compared to expression in AC at 45 DAP. Error bars indicate SD for experiments on three independent fruits.