Figures & data
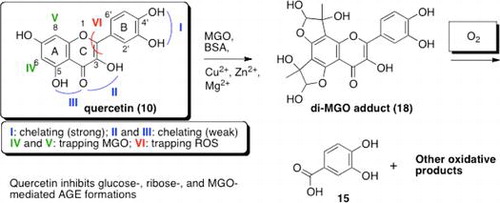
Fig. 1. Effects of metal ions in glucose-BSA, ribose-BSA, and MGO-BSA mediated AGE formation.
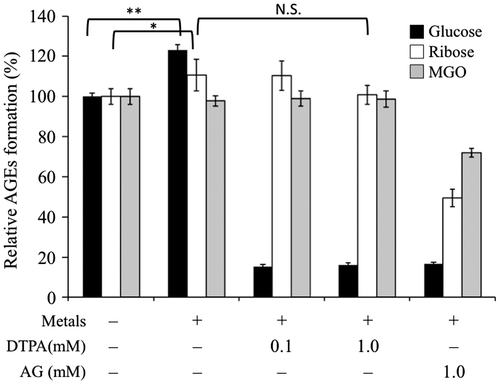
Fig. 2. Inhibitory effects of polyphenols against glucose-BSA, ribose-BSA, and MGO-BSA mediated AGE formation.
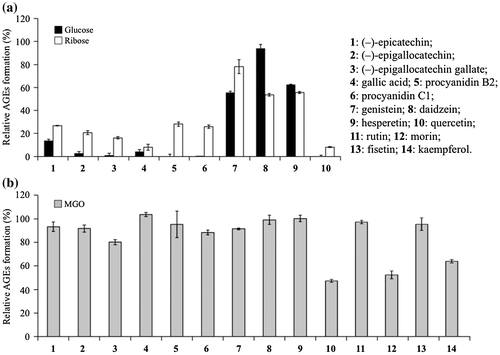
Table 1. 1H and 13C NMR data of isolated 3,4-dihydroxybenzoic acid (15) in MeOH-d4.
Fig. 3. Quantitative analysis of degradation product (15), MGO adducts (16, 17, 18) and quercetin (10).
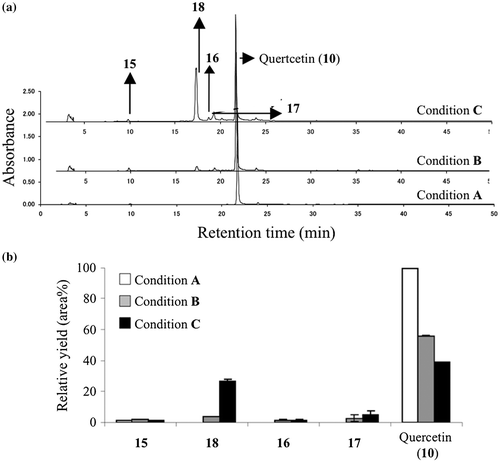
Fig. 4. Proposed mechanism for the fate of quercetin (10) in MGO-mediated AGE formation under ambient air conditions.
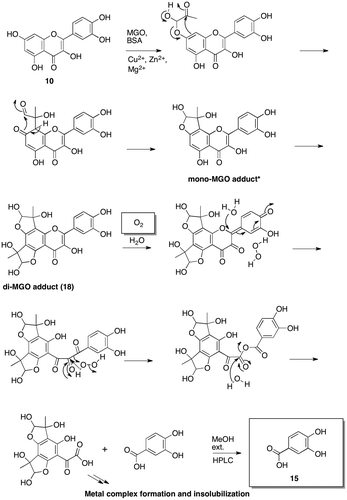
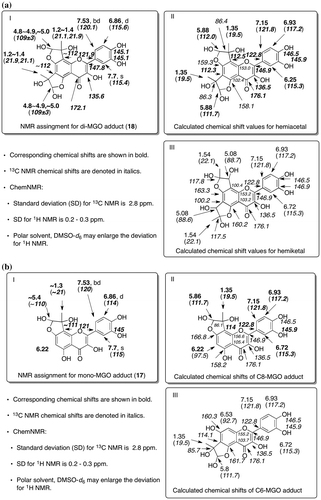
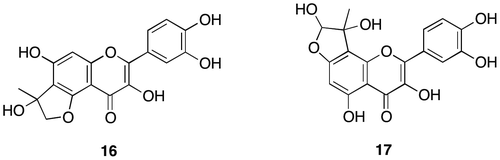
Fig. 6. Structure determination of di-MGO adduct (18) and mono-MGO (17).