Figures & data
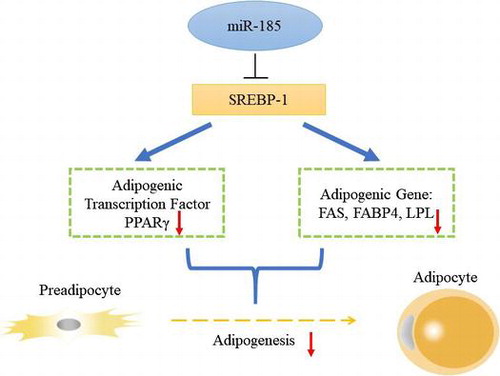
Table 1. The primer sequences used for RT-PCR and gene cloning.
Fig. 1. Expression pattern of miR-185 during 3T3-L1 cell differentiation.
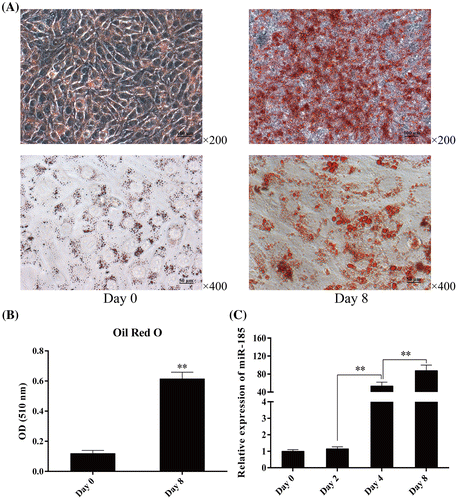
Fig. 2. MiR-185 overexpression inhibits 3T3-L1 cell differentiation.
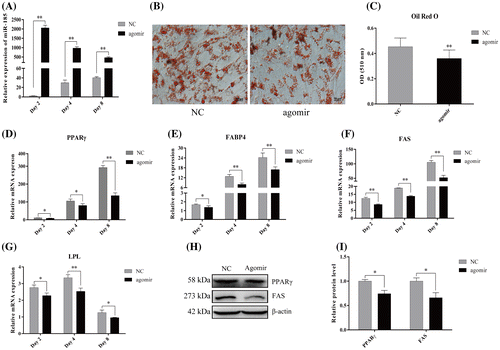
Fig. 3. MiR-185 antagomism promotes 3T3-L1 cells differentiation.
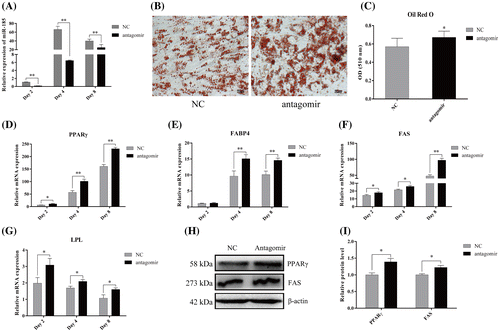
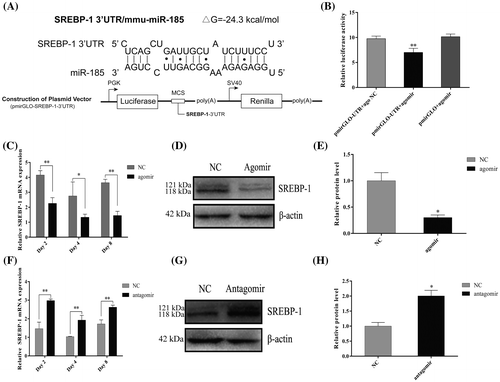
Fig. 4. MiR-185 directly targets the 3′ UTR of SREBP-1 to regulate 3T3-L1 cell differentiation.