Figures & data
Figure 1. GPVI signaling remains constant in platelets spread on fibrous collagen for at least 3 h. (A) Confocal reflection images of washed human platelets spread on Horm collagen for the indicated times. Quantification of spread platelet surface area (B) and platelet number per field of view (C) for the specified time points from 5 FOVs from three independent experiments. Arrows indicate collagen fibers. Scale bar: 5 μm. (D) Western blot analysis of platelets spread for the indicated time points or non-adhered platelets (NA) probed for total phosphotyrosine (4G10), phosphorylated Syk (Tyr525/6; pSyk) and LAT (Tyr200; pLAT). The antibodies against α-tubulin, pan-Syk and pan-LAT were used as loading controls for total phosphotyrosines, phospho-Syk and phospho-LAT, respectively. Quantification of live-cell Ca2+ imaging of platelets spread for the indicated time points showing percentage of platelets exhibiting Ca2+ spiking (E), number of spikes per platelet in the 2 min imaging period (F), spike amplitude (G), spike duration (H). Data taken from three representative FOVs for each time point (~150 platelets in each FOV) from three independent experiments. All data are expressed as mean ± SEM. Significance was calculated using one-way ANOVA with Tukey’s multiple comparisons test (* p < .05)
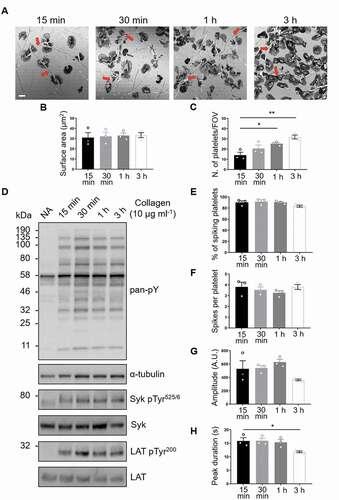
Figure 2. Phosphorylated Syk and LAT colocalize with GPVI along collagen fibers. (A) Confocal microscopy imaging of human washed platelets spread on collagen (10 µg ml−1) for 1 h, labeled for (Ai) Pan-Syk, (Aii) pSyk Tyr525/6, (Aiii) Pan-LAT, (Aiv) pLAT Tyr200. The enrichment of pSyk Tyr525/6 and pLAT Tyr200 along collagen fibers is indicated by red arrows (Aii, iv). (B) Dual-color confocal imaging of platelets, pre-incubated for Pan-GPVI using 1G5-Fab (Bi), spread on collagen for 1 h and post-labeled for pSyk Tyr525/6 (Bii, first panel) or pLAT Tyr200 (Bii, second panel). The position of collagen fibers is shown in the confocal reflection image (Biii). The qualitative colocalisation mask in (Biv) shows the colocalisation of GPVI and the respective phosphoprotein with enrichment at the collagen fibers (arrows). (C) Quantification of GPVI and phosphoprotein colocalisation using Pearson’s correlation coefficient for platelets spreading for 1 h and 3 h. Scatter plot represents mean ± SEM of n = 30 platelets taken from three independent experiments. Significance was measured using unpaired two-tailed t-test, * p < .05. Scale bar: 5 μm
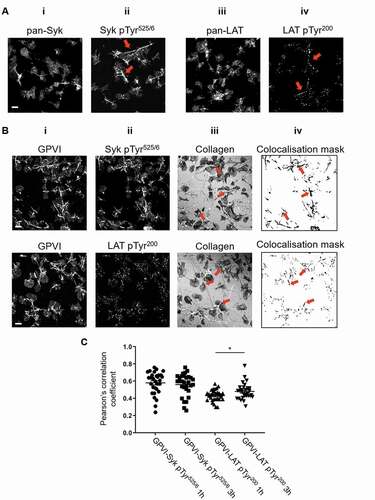
Figure 3. dSTORM imaging and two-level DBSCAN cluster analysis of GPVI in platelets spread on collagen for 1 h and 3 h
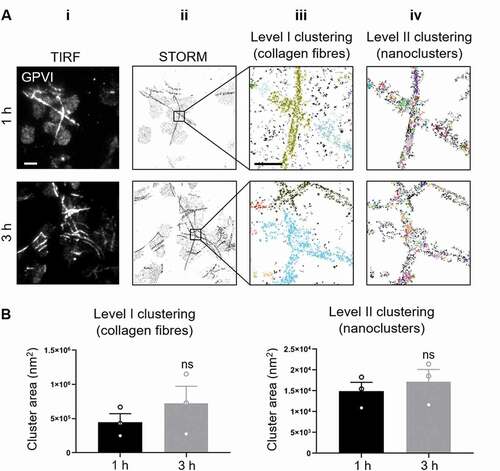
Figure 4. GPVI is not shed in platelets spread on fibrous collagen
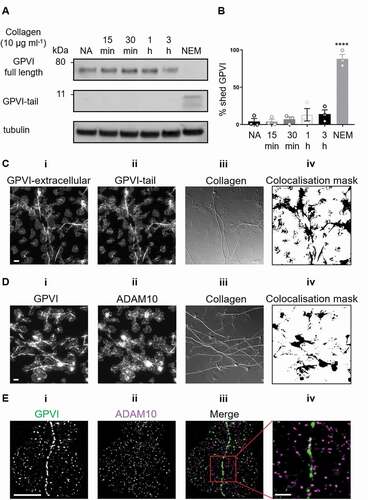
Figure 5. Effect of metalloproteinase inhibitors and NEM on GPVI clustering
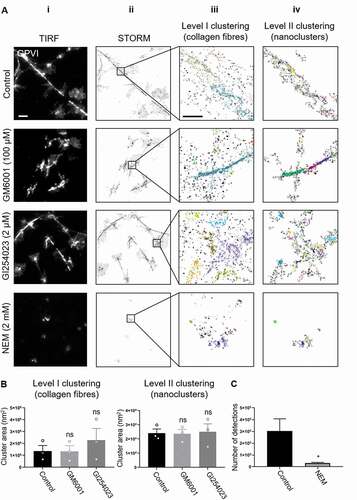
Figure 6. Effect of Src-family and Syk inhibitors on platelet spreading, GPVI signaling and clustering in response to fibrous collagen
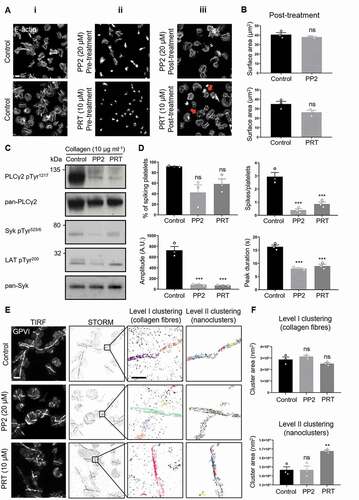
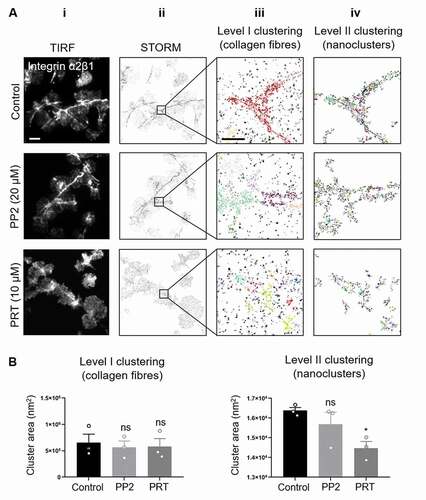
Figure 7. Syk but not Src-family inhibitor impairs integrin α2β1 clustering on collagen in spread platelets