Figures & data
Figure 1. Number of known disease mutations in GPCRs. Data obtained from DisGeNET v 7.0 on all missense variants with a level of evidence greater than or equal to 0.8.
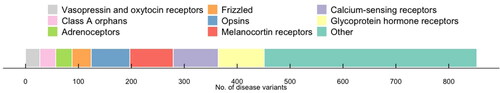
Figure 2. Gain of function (GOF) and loss of function (LOF) mutations in GPCRs. General mechanisms of GOF mutations in GPCRs that include altered receptor selectivity for a specific probe, a gain or change in the preferred signaling pathway triggering constitutive activity, increased binding and/or signaling kinetics, a change in the ligand-modality, as well as increased transportation or recycling may all lead to an amplified receptor activity capacity. On the other hand, LOF mutations in GPCRs may result in an overall disruption or decrease of binding, potency, efficacy, or expression parameters including alterations in receptor stability, folding, trafficking, or the ability to interact with other scaffolding or signaling proteins.
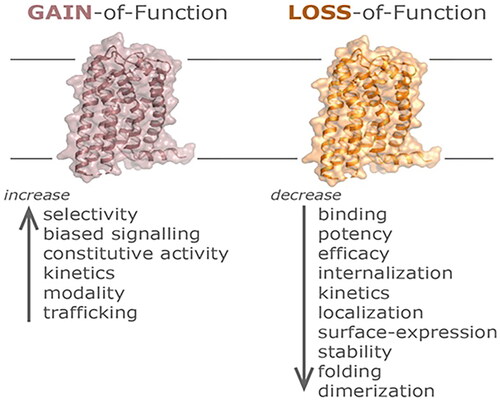
Table 1. Selected G protein-coupled receptors (GPCRs) associated with human genetic disease.
Figure 3. Above, a schematic showing the major structural regions of the CaSR extracellular domain (ECD). the CaSR forms a functional homodimer (protomer 1 in brown and protomer 2 in gray) comprising a bilobed Venus Flytrap domain (VFTD) and a cysteine-domain (CRD). the VFTD contains the ligand-rich binding sites for Ca2+ and amino acids including LR-Trp. Structural studies indicate there are at least two Ca2+-binding sites in each protomer with the cartoon depicting those identified in the recent cryo-EM structures [Citation109]. Loop 1 and 2 reach across the homodimer interface to contact residues on the opposite protomer. Activating mutations have been identified in several residues within these loops. Both inactivating and activating mutations are concentrated in the region between the two lobes of the VFTD, and the CRD homodimer interface. Left, a recent structural representation of the active site (https://www.rcsb.org/structure/7M3F).
![Figure 3. Above, a schematic showing the major structural regions of the CaSR extracellular domain (ECD). the CaSR forms a functional homodimer (protomer 1 in brown and protomer 2 in gray) comprising a bilobed Venus Flytrap domain (VFTD) and a cysteine-domain (CRD). the VFTD contains the ligand-rich binding sites for Ca2+ and amino acids including LR-Trp. Structural studies indicate there are at least two Ca2+-binding sites in each protomer with the cartoon depicting those identified in the recent cryo-EM structures [Citation109]. Loop 1 and 2 reach across the homodimer interface to contact residues on the opposite protomer. Activating mutations have been identified in several residues within these loops. Both inactivating and activating mutations are concentrated in the region between the two lobes of the VFTD, and the CRD homodimer interface. Left, a recent structural representation of the active site (https://www.rcsb.org/structure/7M3F).](/cms/asset/895970c6-5d1e-41df-9c52-e224c632399c/ilab_a_2286606_f0003_c.jpg)
Figure 4. Missense mutations in the CaSR. (A) Schema of the CaSR (which has 1,078 amino acids) showing the relationship between exons 2-7 and the portions of the protein they encode. Exons 2-6 and the beginning of exon 7 encode the ECD comprising ∼610 amino acids, exon 7 encodes the TMD of ∼250 amino acids including membrane-spanning helices TM1-TM7, ECL1-ECL3, ICL1-ICL3, and the ICD of ∼200 amino acids. The locations of missense variants reported in ClinVar and annotated in GnomAD (version 4.0.0, October 2023) as either pathogenic (pink) or variants of unknown significance (orange) are shown above the bars representing exons. It should be noted that many ClinVar variants have not been functionally analysed and may represent benign non-disease causing variants. (B) Schema of the CaSR showing missense mutations that have been functionally characterized and therefore are known pathogenic variants. Multiple mutations have been reported at some of these residues. Please see online version for color figure.
ECD: extracellular domain; ECL: extracellular loop; ICD: intracellular domain; ICL: intracellular loop; TM: transmembrane spanning helix; TMD: transmembrane domain.
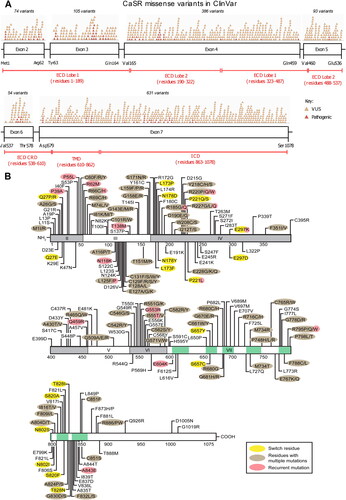
Figure 5. Schematic representation of the V2 receptor and identification of 193 putative disease-causing arginine vasopressin R2 (AVPR2) mutations. Predicted amino acids are shown as the one-letter amino acid code. A solid symbol indicates a codon with a missense or nonsense mutation; a number indicates more than one mutation in the same codon; other types of mutations are not indicated on the figure. There are 95 missense, 18 nonsense, 46 frameshift deletion or insertion, 7 in-frame deletion or insertion, 4 splice-site, 22 large deletion mutations, and 1 complex mutation. The gain of function mutations affecting codons R137 and F229 are indicated. Extracellular, intracellular and transmembrane domains are annotated as reported previously [Citation166].
![Figure 5. Schematic representation of the V2 receptor and identification of 193 putative disease-causing arginine vasopressin R2 (AVPR2) mutations. Predicted amino acids are shown as the one-letter amino acid code. A solid symbol indicates a codon with a missense or nonsense mutation; a number indicates more than one mutation in the same codon; other types of mutations are not indicated on the figure. There are 95 missense, 18 nonsense, 46 frameshift deletion or insertion, 7 in-frame deletion or insertion, 4 splice-site, 22 large deletion mutations, and 1 complex mutation. The gain of function mutations affecting codons R137 and F229 are indicated. Extracellular, intracellular and transmembrane domains are annotated as reported previously [Citation166].](/cms/asset/de6a1fd3-a2ea-4702-87c2-e496534e9e30/ilab_a_2286606_f0005_c.jpg)
