Figures & data
Figure 1. Timeline depicting the year in which antibiotic resistance in each major class of antibiotics was first observed. The main mechanism of resistance for each antibiotic class is also included.
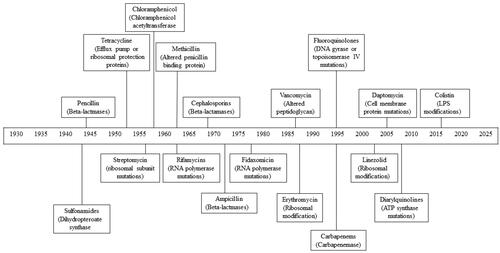
Figure 2. Successive generations of major antibiotic classes over time. Black bonds represent the original skeletal structure while red bonds are structural changes.
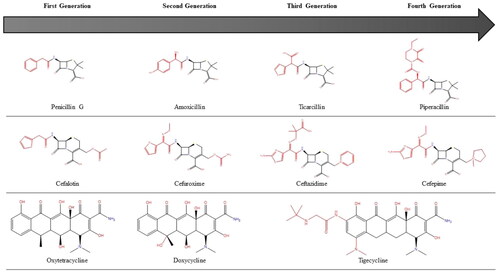
Table 1. Antimicrobial compounds from halophilic and halotolerant actinomycetes.
Table 2. Forward and Reverse primer sequences for Halocins S8, H4 and C8.
Table 3. Characterisation of known halocins, with thermal stability, salt dependency, the spectrum of activity, mechanism, and known sequences.
Table 4. Examples of QSI compounds and their corresponding activity produced from halophilic and halotolerant bacteria.
Table 5. Structures of antimicrobials identified through genome mining from five species of bacteria.
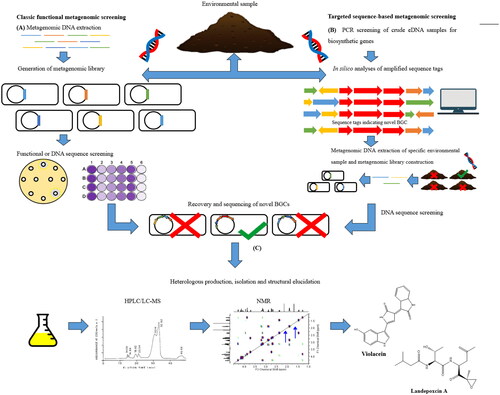
Figure 3. Summary describing the two main approaches that used for antimicrobial discovery from metagenomes. Adapted from Hug et al. (Citation2018).