Figures & data
Table 1. LncRNA classes.
Figure 1. Milestones in lncRNA structural characterization. After the discovery of human XIST and mouse H19 in the early 1990s, it took almost twenty years before systematic structural studies on lncRNAs started. Now, the first studies have been published that characterize full-length, native lncRNA 3D structures. These studies set the ground for future high-resolution investigation on these challenging targets.
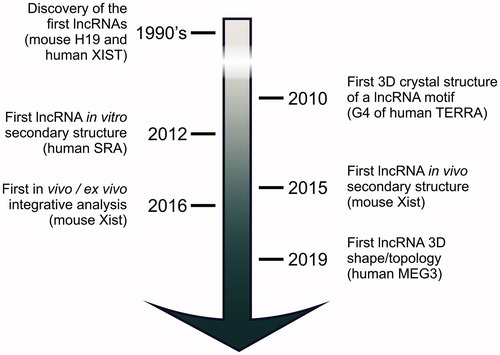
Figure 2. Technology and challenges in lncRNA structural investigation. LncRNA structural studies can be performed by bioinformatics, biochemical assays, secondary and tertiary structure techniques, and functional assays. Each of these approaches have specific informative potential (listed as “information”) and limitations (listed as “challenges”), but when integrated synergistically they can provide a comprehensive molecular characterization of the lncRNA target of interest, and unearth molecular properties that transcriptomic and high-throughput studies would otherwise leave undetected. A color version of this figure is available online.
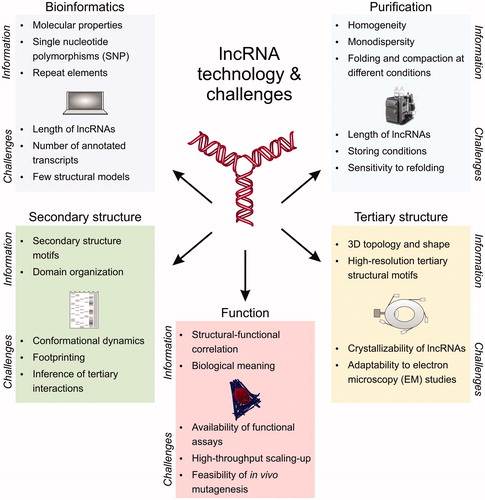
Figure 3. LncRNA structured motifs identified by sequence analysis. Top) RACE sequencing of hLincRNA-p21 revealed that this lncRNA comprises IRAlu elements (left: primary structure). Structure probing of these IRAlus in the context of the full-length hLincRNA-p21 (middle: experimental secondary structure map) surprisingly revealed an architecture similar to that of independently-transcribed Alus, i.e., the Alu of the 7SL subunit of the signal recognition particle, which has been previously crystallized (right: crystal structure from PDB 5AOX). Bottom) Identification of a PAN-like triple helix-forming motif in MALAT1 (left: sequence) fostered the secondary (middle) and high-resolution 3D (right: crystal structure from PDB 4PLX) structure characterization of this short lncRNA motif. A color version of this figure is available online.
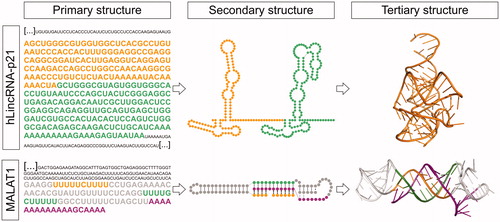
Figure 4. LncRNA structured motifs identified by secondary and tertiary structure analysis. Top) Secondary structure probing of human lncRNA MEG3 (center), revealed the presence of highly conserved intra-molecular interactions, or pseudoknots (complementary sequences highlighted in the left panel), which are essential for conferring MEG3 a surprisingly compact 3D topology (AFM image in the right panel) and its ability to stimulate p53-target gene expression. Bottom) Secondary structure probing of mouse lncRNA Braveheart (middle panel) revealed the presence of a rare RHT motif, named AGIL (sequence highlighted in the left panel). AGIL is crucial for ensuring Braveheart recognition of its partner CNBP, which is the mechanism by which this lncRNA promotes cardiomyocyte differentiation and the correct development of the heart. The 3D structure of Braveheart with CNBP has been studied by SAXS in solution (right panel). MEG3 and Braveheart primary and secondary structures are color coded by exons. A color version of this figure is available online.
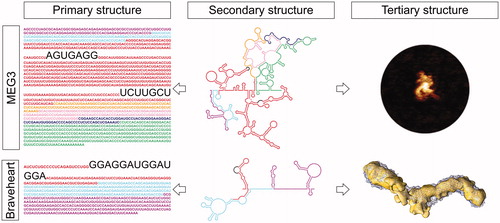
Table 2. Examples of lncRNA secondary structure characterization.
Table 3. Examples of lncRNA tertiary structure characterization.
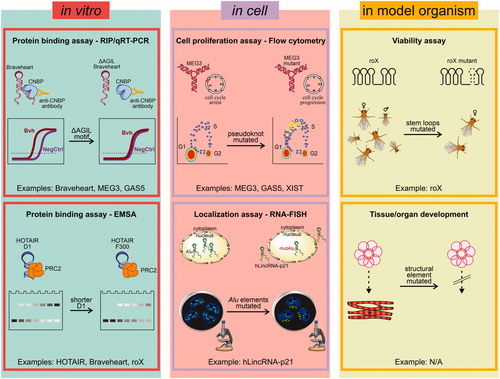
Figure 5. Approaches for functional probing of lncRNA structures. Left) In vitro assays, such as protein binding assays via RIP/qRT-PCR (top) or EMSA (bottom), can be used to test the ability of lncRNA structure mutants to recognize their molecular targets in the test tube. These approaches have been used to probe the structures of Braveheart, MEG3, GAS5, and HOTAIR. Middle) Cell-based assays, such as cell proliferation assays or cellular localization, can be used if lncRNA structural mutants preserve their cellular functions. These approaches have been used to probe the structures of MEG3, GAS5, XIST, and hLincRNA-p21. Right) LncRNA structures can also be probed by genetically engineering structure-based mutations into model organisms and performing viability or developmental assays. These assays are generally more complex and have lower throughput, and have thus so far only been used for roX (N/A = not available). A color version of this figure is available online.