Figures & data
Figure 1. General molecular structure of polyhydroxyalkanoates (Chen & Wu 2005). x=1, 2, 3, yet x = 1 is most common, n can range from 100 to several thousands. R is variable. When x = 1, R = CH3, the monomer structure is 3-hydroxybutyrate, while x = 1 and R = C3H7, it is a 3-hydroxyhexanoate monomer.
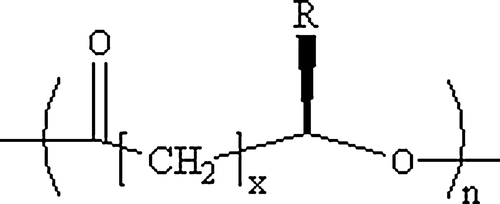
Table 1. Comparison of polymer properties (Khanna & Srivastava, 2005)
Figure 2. Various applications of PHA in medical fields. (A) Artificial esophagus; (B) Vascular patches; (C) Nerve conduit tissue engineering; (D) Articular cartilage tissue engineering; (E) Nutritional and therapeutic uses by oral administration.
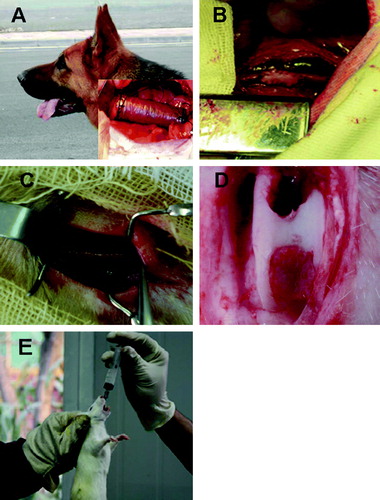
Table 2. Patents for medical application of polyhydroxyalkanoates since 2000
Figure 3. Fibronectin (green) distribution and actin filament formation (red) in SMCs cultured on test films(Qu et al. 2005). (A) SMCs cultured on PHBHHx stained with anti-fibronectin revealed an intense, granular and an oriented delicate fibrillar extracellular labeling pattern. Fibronectin was co-localized with actin filaments clearly (arrow). (B) SMCs on P-PHBHHx stained with anti-fibronectin revealed an intense, perinuclear, granular labeling pattern. Fibronectin was co-localized with actin filaments obviously (arrow). (C) SMCs on Fn-PHBHHx stained with anti-fibronectin revealed a random granular labeling pattern. Fibronectin revealed almost no co-localization with actin filaments (arrow). (D) SMCs cultured on PFn-PHBHHx stained with anti-fibronectin revealed a tense, granular, perinuclear labeling pattern. The co-localization between fibronectin and actin was impaired in some cells (blue arrow) while normal in others (white arrow). (A) PHBHHx; (B) P-PHBHHx; (C) Fn-PHBHHx; (D) PFn-PHBHHx (confocal microscopy, LSM510 meta, Zeiss).
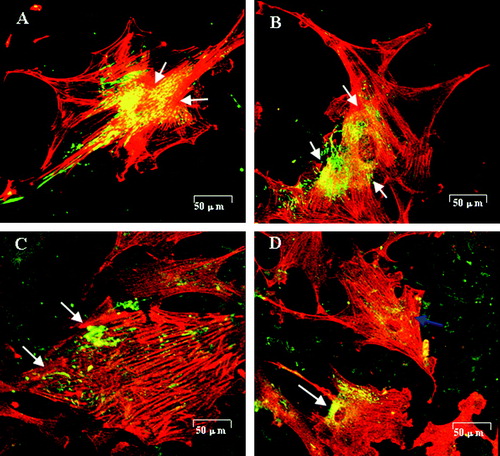
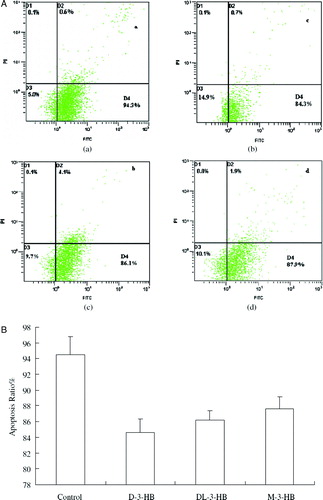
Figure 4. Glial cell apoptosis study by Annexin V-FITC staining assay (Xiao, Zhao, & Chen 2007). Glial cells at an initial concentration of 105 cells/well were grown in DMEM medium supplemented with 20% FBS for 24 h. After the cells attached on the bottom of the wells, the culture medium was replaced by DMEM medium. D-3-HB, DL-3-HB and M-3-HB were all added in 10 mM to the culture medium, respectively. After 48 h, the cells were harvested and stained by Annexin V-FITC and PI. The dead cells were stained by PI and the cells in apotosis were stained by Annexin V-FITC and allocated in the D4 region. (A)—(a) control: cells cultured with DMEM medium; (b) cells cultured with DMEM supplemented with 10mM D-3-HB; (c) cells cultured with DMEM supplemented with 10mM DL-3-HB; (d) cells cultured with DMEM supplemented with 10mM M-3-HB. (B) The percentage of cell apoptosis: statistic result obtained by averaging 6 flow cytometry results of each group in (A).