Figures & data
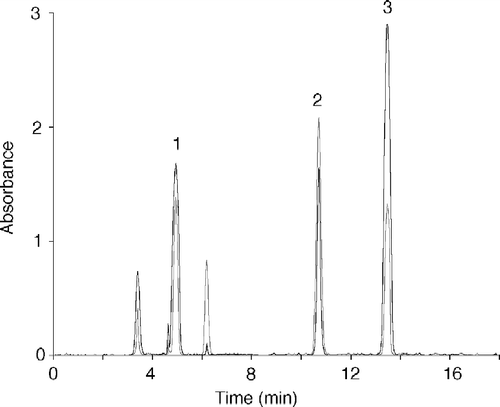
Figure 1 Assays of xanthine oxidase on electrophoretic gels with ethanol, retinol and hypoxanthine via PMS/tetrazolium stain. Zymograms of xanthine oxidase acting on (a) 1 mM ethanol, (b) 1 mM t-retinol and (c) 0.1 mM hypoxanthine are shown. Staining solution composition varied only for the kind of substrate used. Development times were 10 min for hypoxanthine and 1 h for t-retinol and ethanol. The enzyme concentrations assayed with ethanol were decreasing from 2.16 to 1.08 pmoles and those assayed with t-retinol or hypoxanthine were 1.08 and 2.16 pmoles. One representative of three independent experiments is shown.
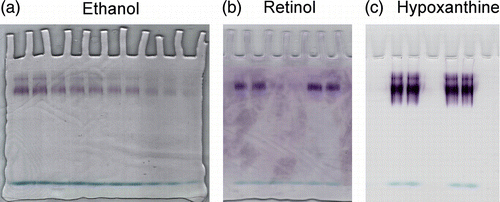
Table I. Kcat values in the conversion of trans-retinol to trans-retinaldehyde and to trans-retinoic acid catalyzed by xanthine oxidase.
Figure 2 Purification of cellular binding proteins from cytosol of human mammary epithelial cells. 100 μL cytosol (0.8 × 106 cells - 74 μg protein) were gel filtered on a KW 804 column and eluted at 1 mL min− 1 with 50 mM Tris HCl pH 7.4 containing 1 mM glutathione. Elution peaks were monitored by absorbance at 280 nm (top trace) and fluorescence (bottom trace). Iterative analyses were carried out and fraction peaks were collected from the area containing cellular retinoid binding proteins (RT 11.47 ± 0.3) (grey peaks). Positions of standards used as molecular weight markers were: 1) bovine erythrocyte carbonic anhydrase (29.3 kDa); 2) horse heart cytochrome C (12.4 kDa).
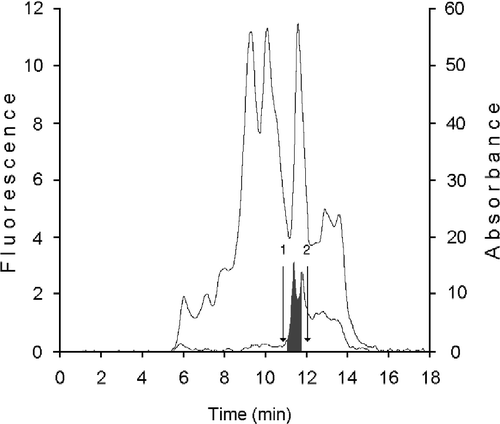
Figure 3 HPLC analysis of cellular retinoid binding proteins from human mammary epithelial cells. CRBP(s) fraction from KW 804 chromatography was analyzed on Protein Pak I-60 column. Bottom trace corresponded to ultraviolet detection at 280 nm; top trace is fluorescence reading (ex. 350 nm; em. 470 nm). CRBPs and CRABPs were separated in the RT interval of 6.4–8.3 min (grey area). Retention times were determined for (1) bovine erythrocytes carbonic anhydrase, 29.3 kDa, (2) lactalbumin, 14.7 kDa, and (3) horse heart cytochrome C, 12.4 kDa.
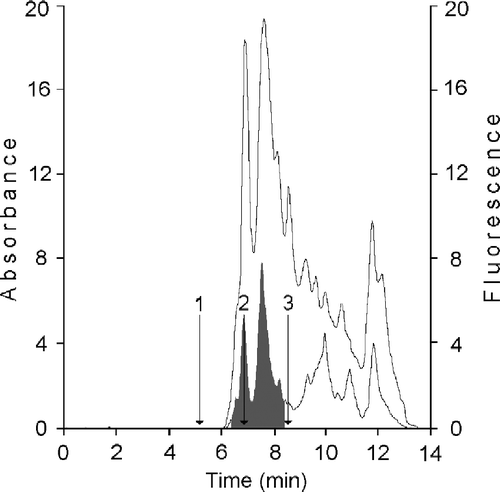
Figure 4 Chromatographic analysis of retinoids produced by xanthine oxidase assayed with t-ROL-CRBP. The chromatographic pattern reported is related to an assay performed with 5 nM xanthine oxidase, 256 nM t-ROL, 25.6 nM CRBP and 250 nM CRABP. Chromatographic elution was performed with acetonitrile-10 mM ammonium acetate (65:35 v/v) at a flow of 1 mL min− 1 and retinoids were monitored at 325 nm (grey profile) and 350 nm (dark profile). Peaks are as follows: 1, all trans-retinoic acid; 2, all trans-retinol; 3, all trans-retinaldehyde.