Figures & data
Figure 1 a) AChE crystal structure (1N5M) with highlighted secondary structure elements and active site entrance. Beta sheets are yellow, alpha helical structures are violet and unordered structures are grey. Catalytic triad is represented by bold tubes. b) A detailed view of the AChE active site, which is located under the omega loop. Secondary structures are highlighted and active site residues are represented by bold tubes. The numbering used is valid for human and mouse structures of AChE. (Please see colour online)
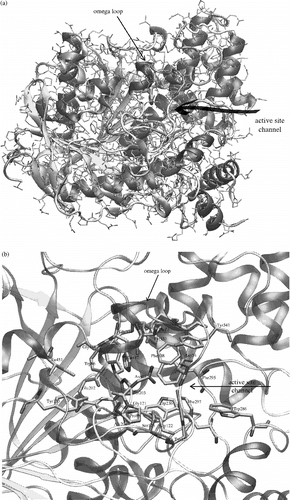
Figure 2 Alignment of primary structures of AChE sequences from Homo sapiens (HUMAN), Mus musculus (MOUSE), Torpedo californica (TORCA) and Drosophila melanogaster (DROSO). Positions of secondary structures depicted under alignment are taken from mouse 1N5M AChE structure. The red arrows point to active site residues. The grey row of bars under each row of alignment is a graphical representation of the alignment score for each column of residues. The score is also marked by signs (star, colon, full stop, space) above the alignment rows. Aminoacid residues are highlighted by colours: acidic (D,E) are red, basic (K,R,H) are blue, polar (N,Q,S,C,T,W,Y) are yellow, non-polar (A,V,L,I,M,F) are green and the others (P,G) stayed white. The far right column shows the number of the last residue in the row. Colour online version. (Please see colour online)
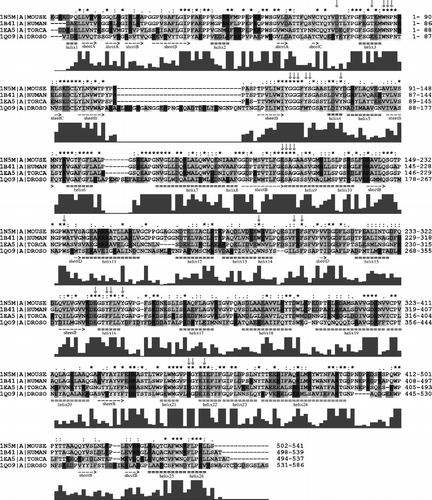
Figure 3 Alignment of primary structures of AChE sequences from Homo sapiens (HUMAN), Mus musculus (MOUSE), Torpedo californica (TORCA). For further description, see Figure 2. For more details see the text. (Please see colour online)
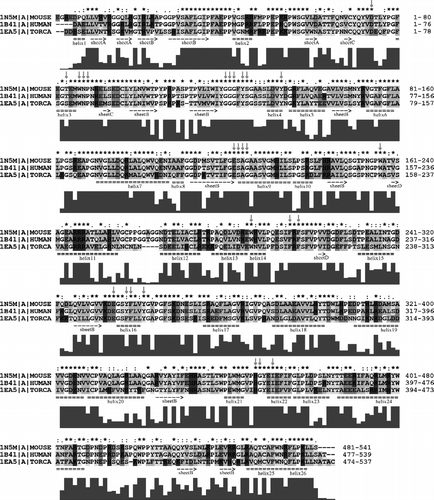
Figure 4 Alignment of primary structures of AChE sequences from Homo sapiens (HUMAN), Mus musculus (MOUSE), Rattus norvegicus (RAT), Felis silvestris catus (FELCA), Oryctolagus cuniculus (RABIT) and Bos taurus (BOVIN). For further description, see Figure 2. For more details see the text.
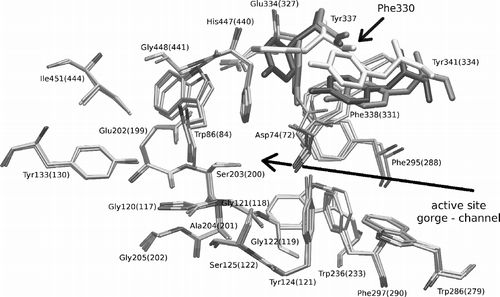
Figure 5 3-D alignment of human 1B41 and drosophila 1QO9 structures. Mutated residues and flipped Trp86(83) are coloured by red for human and orange for the drosophila structure. The first residue number always belongs to the human AChE and “/” character means a mutation, where the first aminoacid is from the human structure. Colour online version.
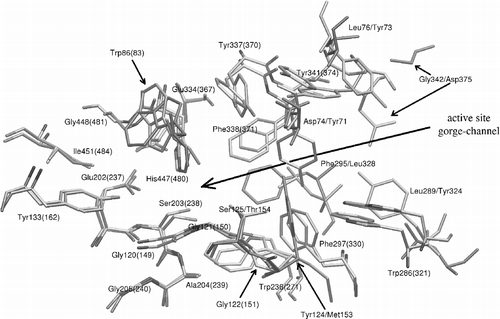
Figure 6 3-D alignment of human 1B41, mouse 1N5M and torpedo 1EA5 structures. Residues which show the biggest deviations are coloured by red for human, blue for mouse and yellow for the torpedo structure. Torpedo californica has also one mutation of Tyr337 (in the case of human and mouse) to Phe330 (torpedo). The first residue number belongs to the human or mouse AChE. Colour online version.