Figures & data
Table I. Data set used for CoMFA analysis with Ki (nM) and pKi values in the quadruple mutant (Asn51Ile, Cys59Arg, Ser108Asn, Ile164Leu) of PfDHFR.
Figure 2. The backbone superimposition between the X-ray PfDHFR/WR99210 complex structures of quadruple mutant (PDB entry 1J3K shown in green color) and PfDHFR/pyr double mutant (PDB entry 1J3J shown in orange color), indicating the similarity binding position of both of the inhibitors.
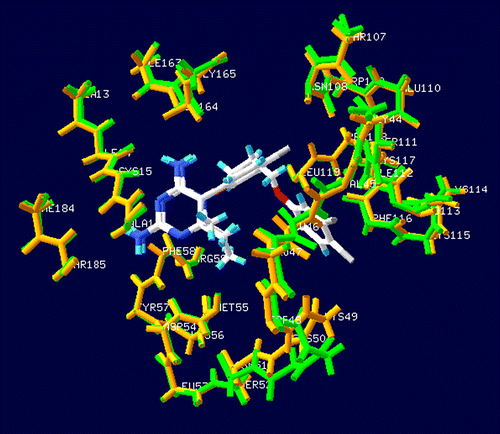
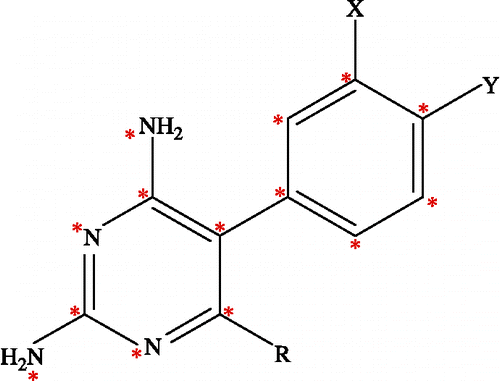
Figure 3. The 2D scheme of the adopted model system of inhibitor bound to the quadruple mutant type (Asn51Ile, Cys59Arg, Ser108Asn, Ile164Leu) of PfDHFR binding site.
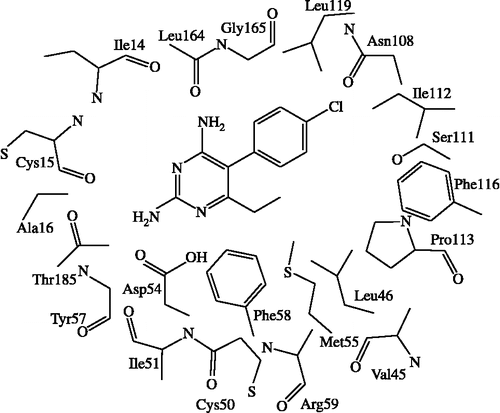
Table II. PLS statistical results of CoMFA models for quadruple mutant PfDHFR.
Table III. Actual (Act) and predicted (Pred) pKi values and the residuals (Δ) of the training set and test set molecules for the mutant PfDHFR models.
Figure 4. Plot of the predicted and actual pKi values of the training set and the test set molecules with CoMFA H model IV.
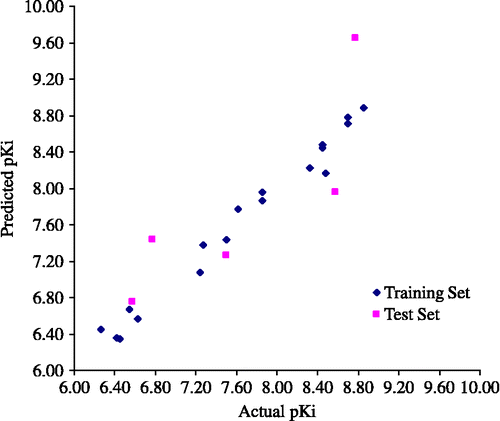
Figure 5. CoMFA (stdev.*coeff.) sterically favored areas are represented by green regions. Sterically unfavored areas are represented by yellow regions (level of steric contour contribution = 80%) and compound 12 is represented by ball and stick.
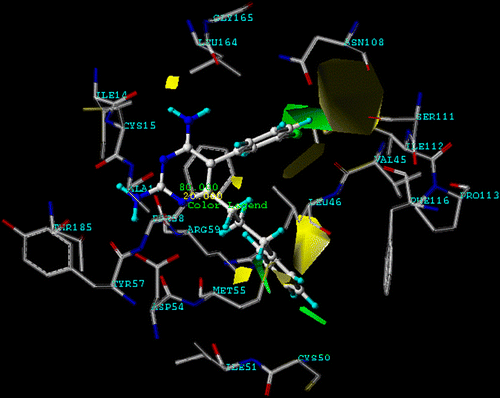
Figure 6. CoMFA (stdev.*coeff.) negative charge favored area is represented by the red region. Positive charge favored area is represented by the blue region (level of electrostatic contour contribution = 80%) and compound 12 is represented by ball and stick.
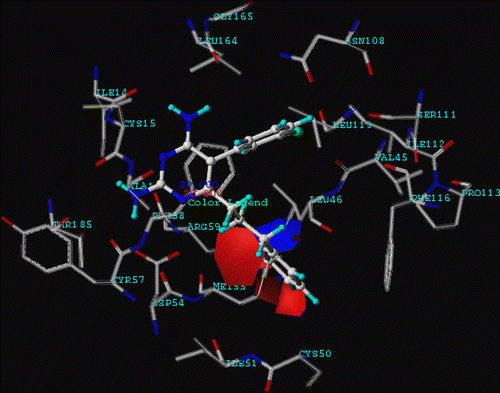
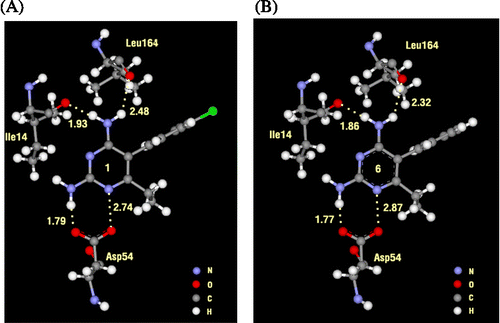
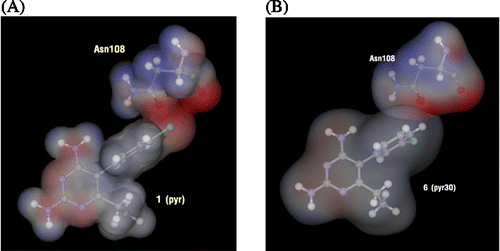
Table IV. Particular interaction energy (kcal/mol) of 1 (pyr) and 6 with individual residues, calculated at MP2/6-31G(d,p) and MP2/6-31G(d,p) levels with BSSE-CP.