Figures & data
Figure 2. Concentration-response curves for the effects of cubebin and derivatives on state 3 respiration rate of isolated Hamster liver mitochondria (1.5 mg protein), incubated at 30°C with 2.5 mM glutamate + 2.5 mM malate, in a standard medium containing 125 mM sucrose, 65 mM KCl and 10 mM HEPES-KOH, pH 7.4, in presence of 0.5 mM EGTA and 10 mM K2HPO4, in a final volume of 1.8 mL. State 3 respiration was initiated with 720 nmol ADP. Data are mean ± SD of three independent experiments. Percent of inhibition in relation to control: 57.84 ± 14.69 ng atoms O/min/mg protein. Respiratory control ratio: 4.40 ± 1.13; ADP/O: 1.41 ± 0.08.
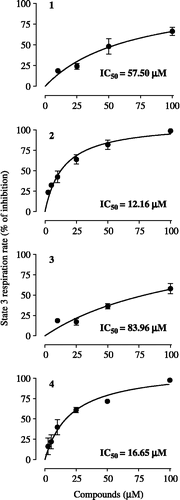
Figure 3. Concentration-response curves for the effects of cubebin and derivatives on submitochondrial particles (10 μg protein) NADH oxidase activity employing 100 μM NADH as substrate, at 30°C, in the standard medium described in legend of Figure 2, in presence of 0.2 mM EGTA (1 mL final volume). Data are mean ± SD of three independent experiments. Percent of inhibition in relation to control: V0 = 0.00637 ± 0.002 μmol/min/mg protein.
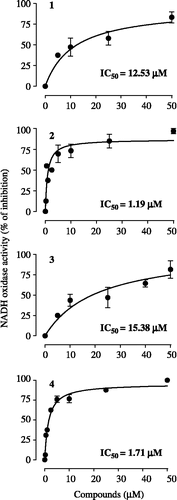
Figure 4. Double-reciprocal plots for the inhibition of submitochondrial particles NADH oxidase activity by cubebin and derivatives, assayed as described in legend of . Data are mean ± SD of three independent experiments. (○) absence of cubebin/derivatives; (•) presence of cubebin/derivatives in concentrations corresponding to the respective IC50 values, given on . Km and Vmax for control: 27.65 μM and 0.00733 μmol/min/mg protein, respectively. KI: inhibition constant.
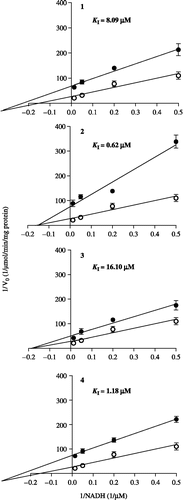
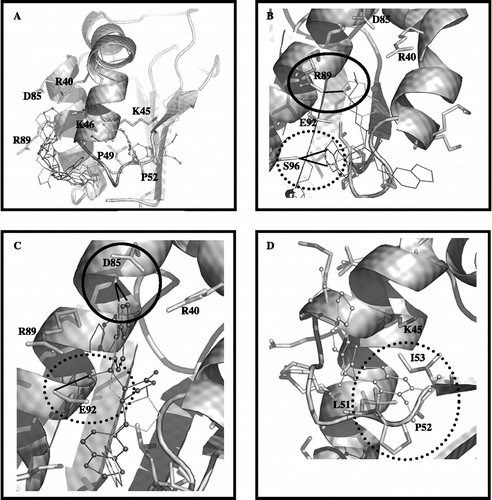
Figure 5. In (A), the top-ranked solution for flexible docking between the compounds/rotenone with CI-B8. The residues having the strongest interaction with the inhibitors, are shown. In (B), the superposition between the top-ranked solutions for the most active compounds 2. (in light grey) and 4 (in dark grey). The circle demonstrates the possible hydrogen bond between residue R89 and oxygen of benzodioxol ring of compound 2; the dotted line circle demonstrates hydrogen bond donate by the S96 side chain to carboxyl oxygen of compound 2 or 4. In (C), the superposition between the top-ranked solutions for the less active compounds 1 (in white) and 3 (in black ball line). The circle demonstrates the possible contact between residue D85 and benzodioxol ring of compounds 1 and 3; the dotted line circle demonstrates possible hydrogen bond between residue E92 and hydroxyl group at C7′ of compound 3. In (D), the rotenone orientation; the dotted line circle demonstrates the possible interaction of rotenone with two conserved residues (P52 and I53).