Figures & data
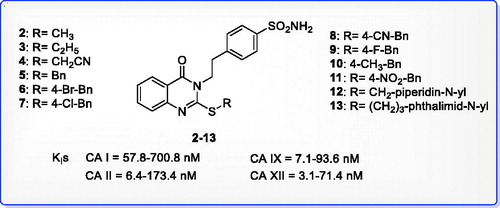
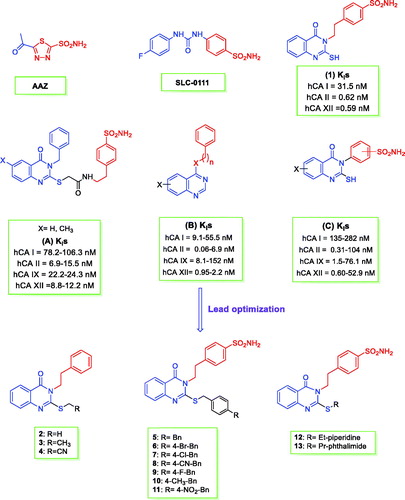
Figure 1. Structures of AAZ, SLC-0111, (A–C), and the herein designed quinazoline derivatives (2–13) as CAIs.
Scheme 1. Synthesis of 4–(2–(2-(substituted-thio)-4-oxoquinazolin-3(4H)-yl)ethyl)benzenesulfonamides 2–13.
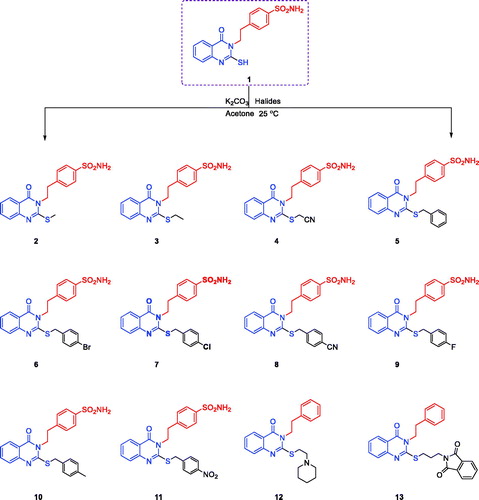
Figure 2. Carbonic anhydrase inhibition of 2-substituted-mercapto-4(3H|)-quinazolinone derivatives 2–13.
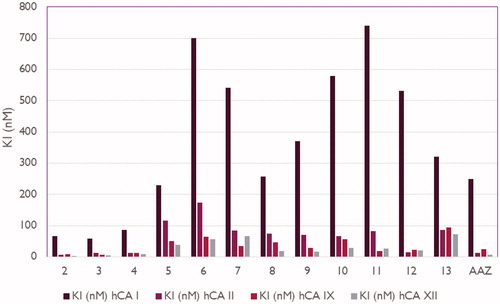
Table 1. Inhibition constant values of 2-ethylquinazoline derivatives 2–13 and standard sulphonamide inhibitor acetazolamide (AAZ) against human CA isoforms hCA I, II, IX, and XII as determined by a stopped flow, CO2 hydrase assay48.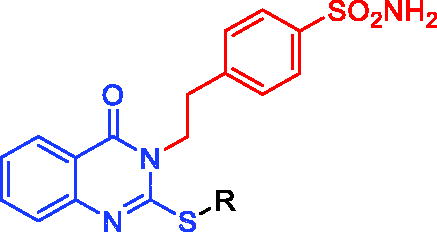
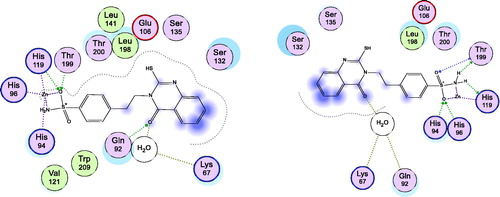
Figure 3. Docking modes of compound 2 in the binding pockets of CA isoenzymes II and XII. Interactions between the protein (PDB IDs: 5ULN and 1JD0). Predicted binding modes of co-crystallized inhibitor (upper left panel) and compound 2 (upper right panel) with hCA-II target as well as co-crystallized inhibitor (lower left panel) and compound 2 (lower right panel) with hCA-XII target.
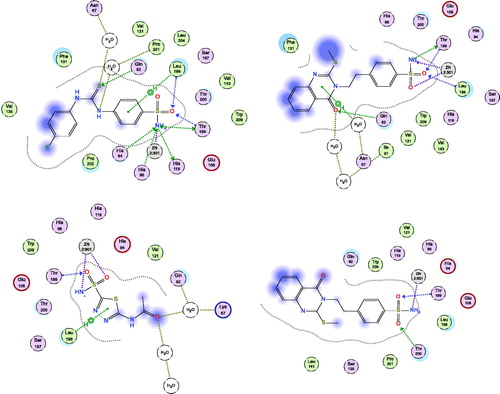
Figure 4. Docking modes of compound 3 in the binding pockets of CA isoenzymes I and IX. Interactions between the protein (PDB IDs: 4WR7, 5FL4). Predicted binding modes of co-crystallized inhibitor (upper left panel) and compound 3 (upper right panel) with hCA-I target as well as co-crystallized inhibitor (lower left panel) and compound 3 (lower right panel) with hCA-IX target.
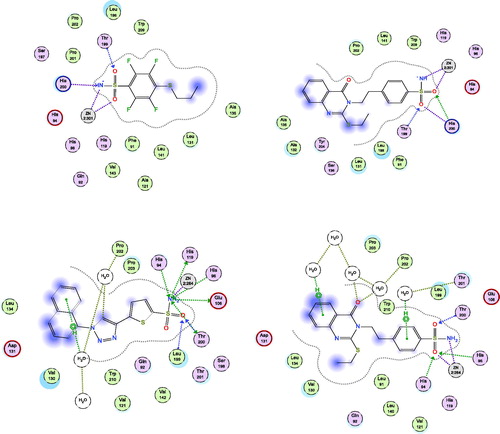
Figure 5. Docking modes of compound 6 as the least active example in the binding pockets of CA isoenzymes I and II. Interactions between the protein (PDB IDs: 4WR7, 5ULN). Predicted binding modes of compound 6 with CA-I; right panel, and CA-II; left panel.
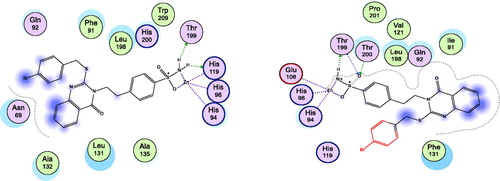
Table 2. Description of the docking data of selected target compounds 2 and 3.