Figures & data
Figure 1. Human topoisomerase Iiα inhibitors based on thiosemicarbazide coreCitation6–8.
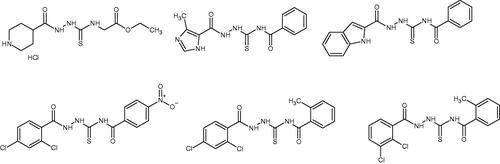
Table 1. Chemical structures of compounds characterised by the most potent affinity towards human DNA topoisomerase II.
Table 2. Inhibition of human DNA topoisomerase Iiα by compounds 1-3 and etoposide.
Table 3. Antiproliferative activity of compounds 1-3 and etoposide examined in BrdU assay.
Table 4. Cytotoxicity of the investigated compounds 1-10 and etoposide against a panel of cancer cell lines measured by using MTT assay after 24 and 48 h incubation.
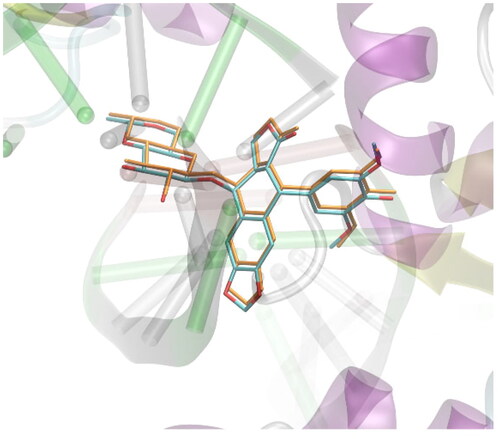
Figure 2. The exemplary, representative poses of the three ligands interacting with binding cavity of topoisomerase II. All depicted poses, characteristic of a given ligand, exhibit similar level of binding energies (see discussion in the text). (A) compound 1; (B) compound 2; (C) compound 3.
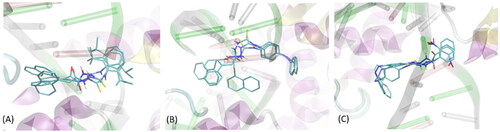
Figure 3. The exemplary, energetically-favourable poses of the three lead compounds identified during docking study: (A) compound 1; (B) compound 2; (C) compound 3.The ligand molecules are shown as thick sticks whereas all the closest amino-acid residues (within the distance of 0.4 nm) are represented by thin sticks. The description of the interaction types is given in the text.
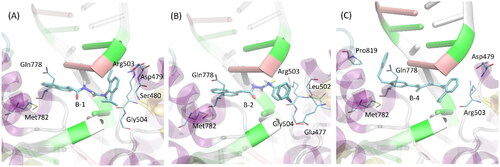
Table 5. Inhibitory effect of compounds 1-3 against indoleamine-2,3-dioxygenase 1 (IDO 1).