Figures & data
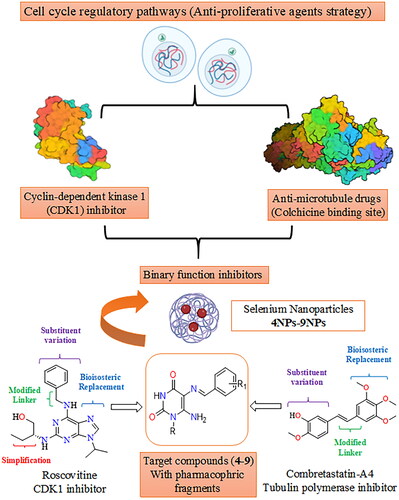
Figure 2. Design of novel pyrimidine Schiff bases and their SeNPs as dual CDK1 and tubulin polymerase inhibitors.
Scheme 1. Formation of Schiff′s bases 4–9. Reagents and conditions: (i) NaNO2/AcOH/H2O, r.t, 30 min; (ii) (NH4)2S, 75 °C, 15 min and (iii) aromatic aldehydes/gl. AcOH/heated under fusion, 20 min.
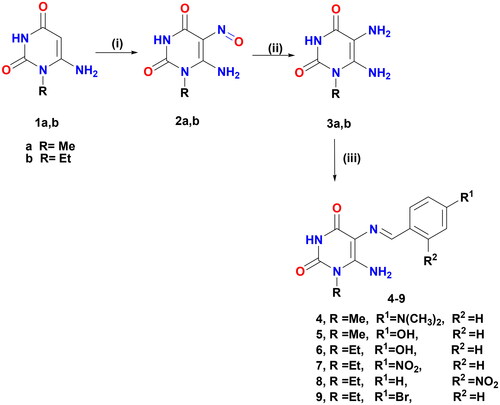
Figure 4. (a) A schematic diagram of the synthesis of selenium nanoparticles; (b) UV-Vis spectrum of selenium nanoparticles.
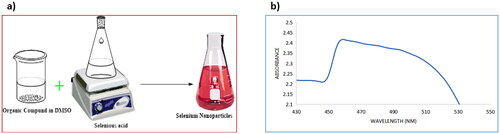
Figure 5. TEM of SeNPs, (a), (b), (c), (d), (e), and (f) TEM of SeNPs loaded onto 4–9, respectively.
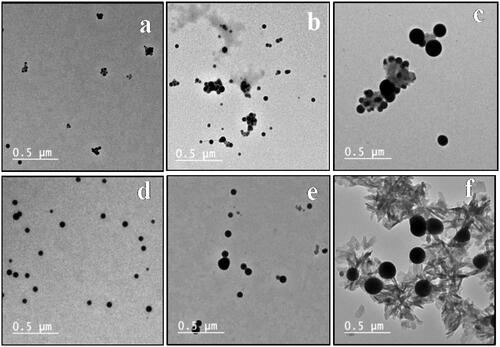
Table 1. Particle size and zeta potential of Het-SeNPs 4NPs–9NPs.
Table 2. The anti-proliferative activity of pyrimidine Schiff bases (4–9) and their nano-sized forms (4NPs–9NPs) against human cancer cell lines (IC50 µM).
Table 3. The selectivity of the most potent candidates 4 and 6 in both normal and nanoformulations (4NPs and 6NPs).
Figure 6. The inhibitory effect of the compounds 4 and 4NPs on (a) cyclin-dependent kinase 1, (b) Tubulin polymerisation through colchicine binding site. (*) indicate to the significant differences between compound 4NPs-treated and compound 4-treated cells, where (***) indicates to p < 0.001. All experiments were performed in triplicates.
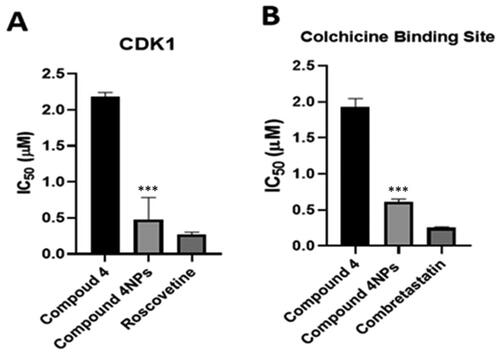
Table 4. The inhibitory activity of the most potent candidates 4 and 4NPs against both CDK1 and tubulin polymerase (CBS) compared to two reference drugs.
Figure 7. (a) The effect of the most potent compounds 4 and 4NPs on cell cycle phases of HepG-2. Flow cytometric histograms of HepG-2 cell cycle phases; (b) untreated cells, (c) treated with compound 4 alone, and (d) treated with compound 4NPs. All experiments were performed in triplicate.
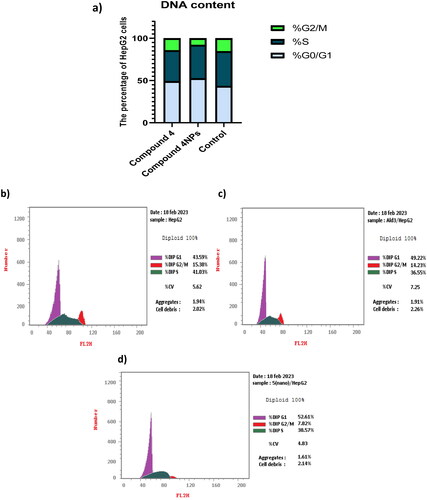
Table 5. The effect of the most potent candidates 4 and 4NPs on cell cycle phases of HepG-2.
Figure 8. (a) The apoptosis-inducing effect of the most potent candidates 4 and 4NPs on HepG-2. (*) indicate to the significant differences between compound 4NPs-treated and compound 4-treated cells, where (*) indicates to p < 0.05, (**) p < 0.01. All experiments were performed in triplicates. (b–d). Flow cytometric dot plot of PI/annexin V screening of HepG-2; (b) untreated cells, (c) cells treated with compound 4, and (d) cells treated with compound 4NPs. All experiments were performed in triplicate.
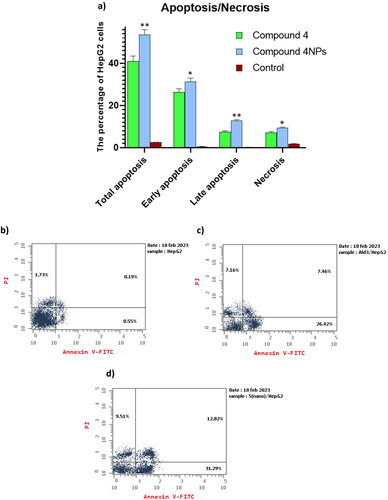
Table 6. The apoptotic effect of the most potent candidates 4 and 4NPs on HepG-2.
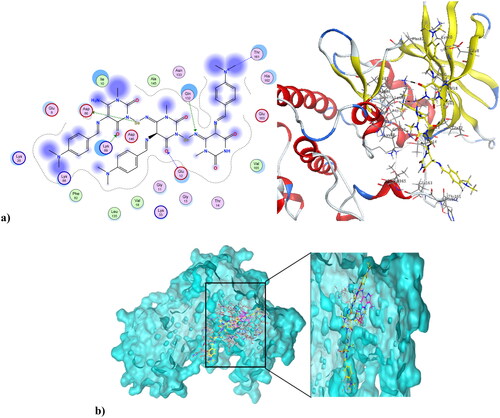
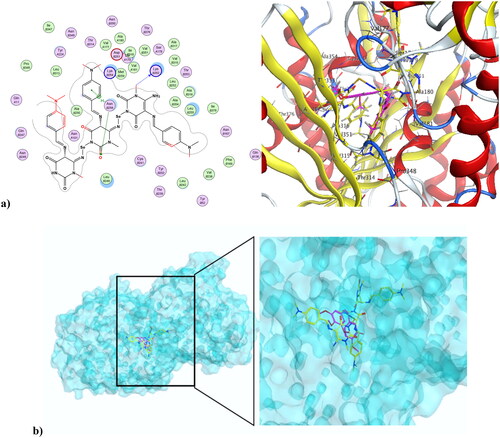
Figure 9. (a) Compound 4 at the active site of CDK1 (PDB ID: 6GU6), and (b) alignment of compound 4 and dinaciclib at the active site of CDK1 (PDB ID: 6GU6).
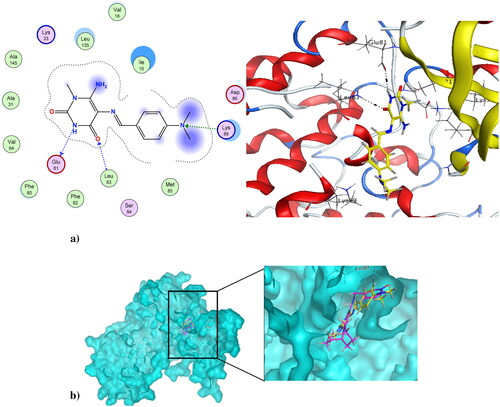
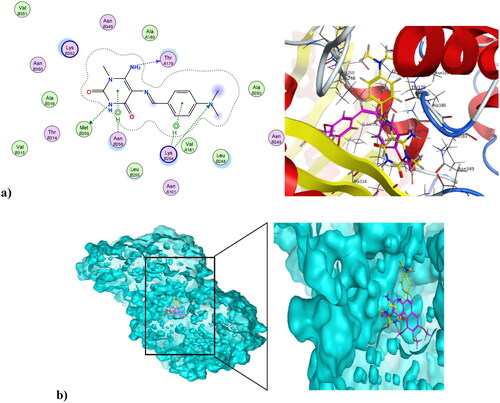
Figure 10. (a) Nano compound 4NPs at the active site of CDK1 (PDB ID: 6GU6). (b) The alignment of 4NPs and dinaciclib at the active site of CDK1 (PDB ID: 6GU6).