Figures & data
Figure 1. The strategy for the development of SARS-CoV-2 S protein-human ACE2 interaction inhibitors based on peptide foldamers.
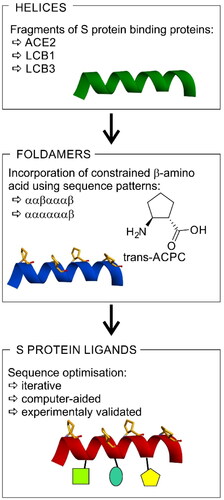
Figure 2. Crystal structure of LCB3-RBD complex (A), PDB id 7JZM (residues 1–18 of LCB3 are shown), and the modelled foldameric peptide 10 bound to the RBD of S protein (B). Peptides are shown in stick representation; trans-ACPC residues’ carbon atoms are coloured green, tryptophan residues’ carbon atoms are coloured pink. The surface of the RBD is coloured according to interpolated charge: blue – positive, grey – neutral, red – negative. Key intermolecular interactions are shown as dashed lines: green – hydrogen bonds, orange – charge assisted hydrogen bonds, pink – hydrophobic interactions, white – hydrogen bond donor/π interactions.
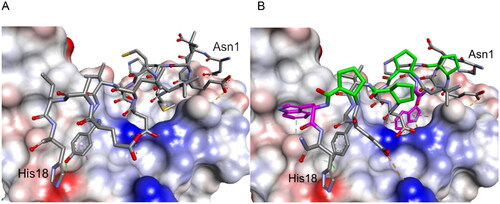
Figure 3. CD spectra of (A) peptides (2–5) derived from LCB1 helix (1) and (B) peptides (6–10) derived from LCB3 helix (6), dissolved in potassium phosphate buffer, 50 mM, pH 7.5.
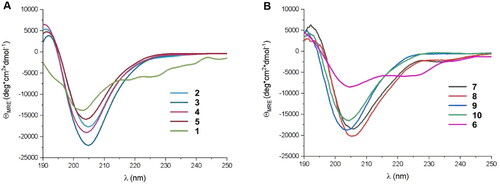
Table 1. Amino acid sequences of the first set of designed helical foldamers (peptides 2–5 and 7–10) and their affinities to the RBD of the S protein.
Figure 4. BLI-based screening of the binding affinity properties of peptide 10 towards RBD. Association and dissociation steps are given for different concentrations of peptide 10, ranging from 1 to 100 µM.
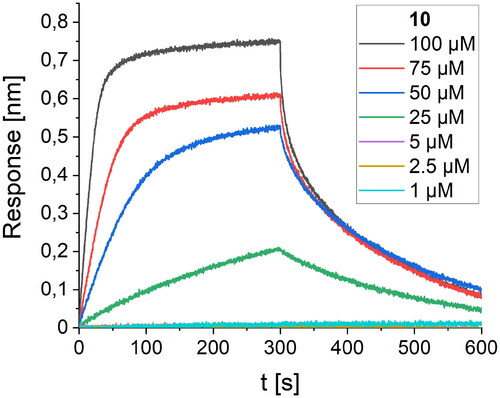
Figure 5. The modelled complex of peptide 12 with the RDB of the S protein. Peptide 12 is shown in stick representation; trans-ACPC residues’ carbon atoms are coloured green, mutated residues’ carbon atoms are coloured pink. The surface of the RBD is coloured according to interpolated charge: blue – positive, grey – neutral, red – negative. Key intermolecular interactions are shown as dashed lines: green – hydrogen bonds, orange – charge assisted hydrogen bonds, pink – hydrophobic interactions, white – hydrogen bond donor/π interactions.
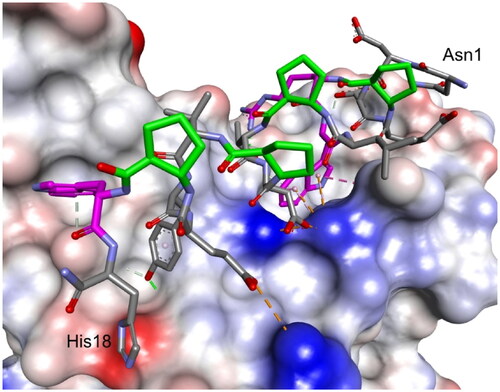
Figure 6. BLI-based screening of the binding affinity properties of peptide 12 towards RBD. Association and dissociation steps are given for the different concentrations of peptide 12, ranging from 1 to 100 µM.
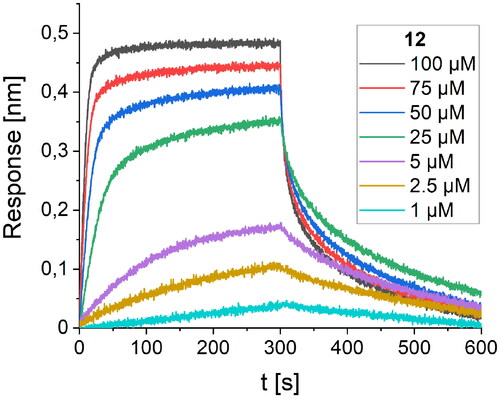
Table 2. Amino acid sequences of the second set of designed helical foldamers (11–18) derived from peptide 10 and their binding affinities to the RBD of the S protein.
Figure 7. BLI-based screening of the binding affinity properties of peptide 27 towards RBD. Association and dissociation steps are given for different concentrations of peptide 27, ranging from 0.25 to 10 µM.
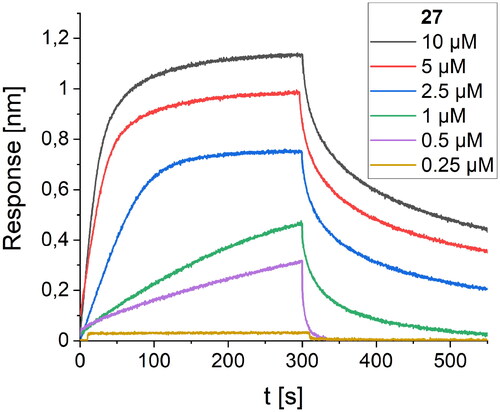
Figure 8. Modelled complex of peptide 27 with the RBD of the S protein. Peptide 27 is shown in stick representation; trans-ACPC residues’ carbon atoms are coloured green, mutated residues’ carbon atoms are coloured pink. The surface of the RBD is coloured according to interpolated charge: blue – positive, grey – neutral, red – negative. Key intermolecular interactions are shown as dashed lines: green – hydrogen bonds, orange – charge assisted hydrogen bonds, pink – hydrophobic interactions, white – hydrogen bond donor/π interactions.
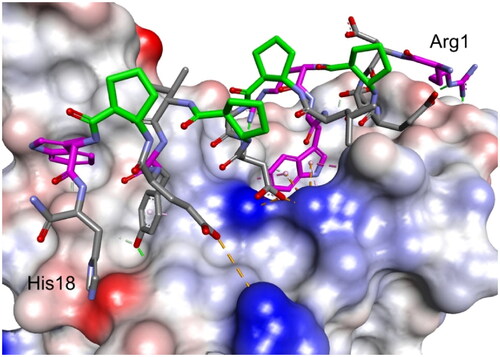
Table 3. Amino acid sequences of the third set of designed helical foldamers (19–32) derived from peptide 12 and their binding affinities to the RBD of the S protein.
Figure 10. BLI-based screening of the binding affinity properties of peptide 41 towards RBD. Association and dissociation steps are given for different concentrations of peptide 41, ranging from 0.25 to 10 µM.
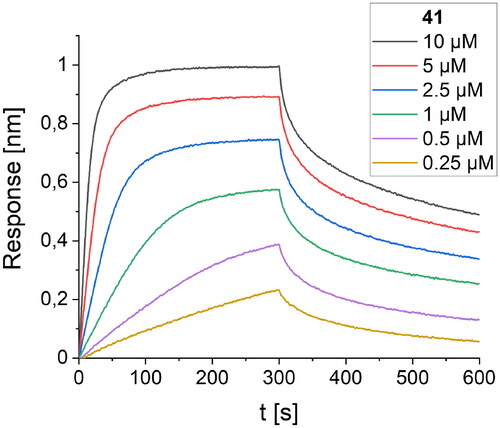
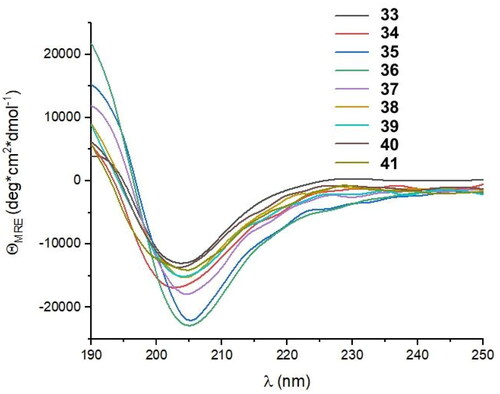
Table 4. Amino acid sequences of the fourth set of designed helical foldamers (33–41) derived from peptide 27 and their binding affinities to the RBD of the S protein.
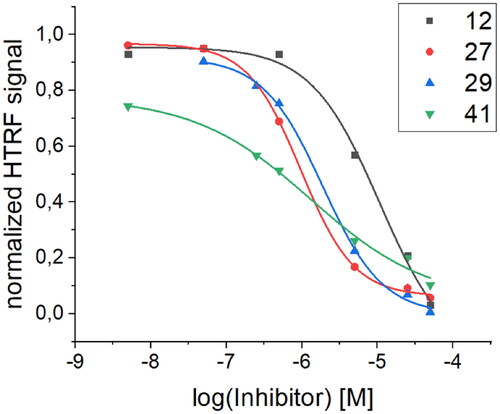
Table 5. Comparison of Kd values obtained via BLI screening and IC50 values obtained from the HTRF assay for selected peptides with the most pronounced activity.
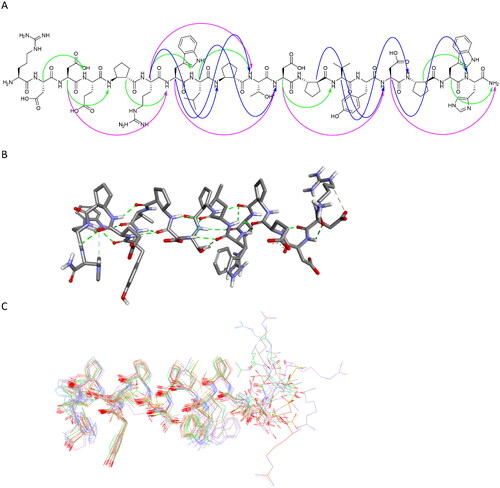
Figure 12. Regular NOESY contacts between HA and HN atoms of nonadjacent residues of peptide 27 (A). Contacts between i - i + 2, i - i + 3 and i - i + 4 are shown in green, blue and pink, respectively. Averaged structure of peptide 27 calculated on the basis of NMR-derived restraints shown in stick representation (B) and superimposition of 10 lowest energy structures (C). Green dotted lines represent hydrogen bonds.
Supplemental Material
Download PDF (2.4 MB)Data availability statement
Data will be made available on request.