Figures & data
Scheme 1. Synthesis of the target compounds. (a) (i) Et3N, MsCl, DCM, 0 °C, 1 h; (ii) NaOH, H2O, 0 °C, 4 h; (b) NH4Cl, NaN3, DMF, 80 °C, 10 h; (c) Pd/C, H2, MeOH, r.t., 8 h; (d) Substituted propionic acid, PyBOP, DIEA, DMF, r.t., 5 h.
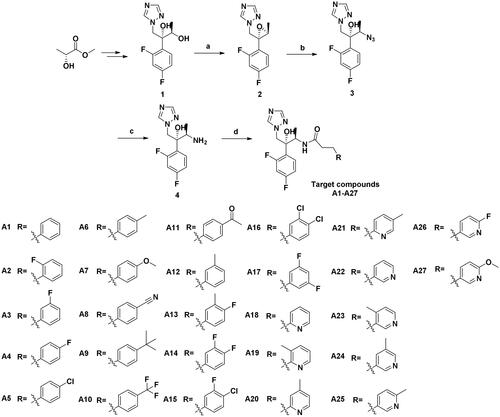
Table 1. In vitro antifungal activity of the target compounds against tested fungi (MIC, μg/mL).
Table 2. In vitro antifungal activity of the potent compounds against FCZ-resistant C. albicans and C. auris isolates measured by MIC (μg/mL).
Figure 4. GC-MS analysis of sterols in C. albicans cells. The fungal strain was treated with DMSO (Control), FCZ, compounds A1, A3 or A9 at 8 μg/mL for 8 h.
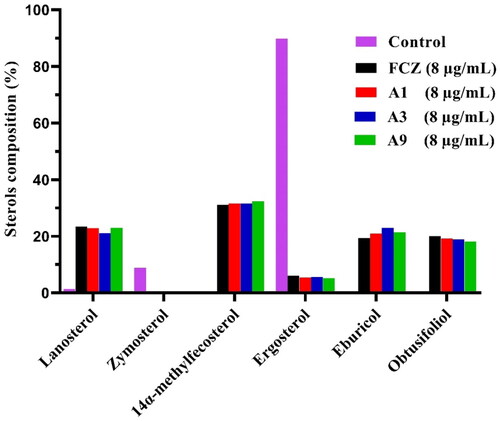
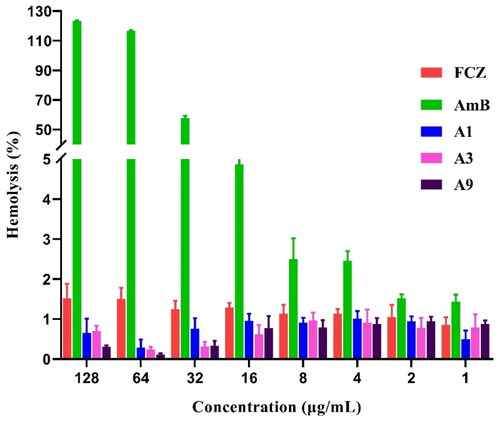
Figure 5. Haemolytic effect of A1, A3 and A9 against rabbit red blood cells at different indicated concentrations.