Figures & data
Figure 1. Left. Reaction of the 1,5-diaryl-3-oxo-1,4-pentadienyl pharmacophore with thiol group at the catalytic site (showing Cys111 and His272) of SARS-CoV-2 PLpro. Initially, a non-covalent enzyme-inhibitor association is formed (E:I, 1) leading to a covalent adduct. Right. Structure of BBC DU-UC15 (2c) with the 1,5-diaryl-3-oxo-1,4-pentadienyl pharmacophore in red.
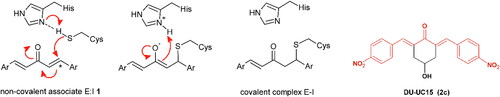
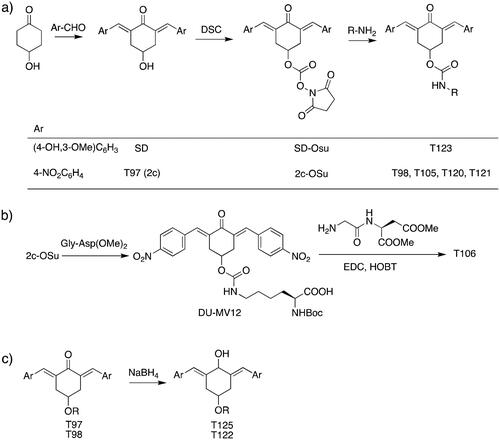
Table 1. Bis(benzylidene)cyclohexanones studied in this work.
Table 2. Chemical structures of the reference inhibitors of Mpro and PLpro of SARS-CoV-2 or proteasome-associated deubiquitinating enzyme inhibitors included in this study.
Figure 2. Left: 3D structure of the SARS-CoV-2 PLpro-GRL0617 complex (PDB entry 7CMDCitation62). The inhibitor GRL0617 that occupies the S4 - S3 pockets of the substrate binding cleft is shown in the CPK representation. Catalytic triad of PLpro: Cys111 - His272 - Asp286 coloured purple shows the location of the catalytic site. Right: 2D scheme of inhibitor - residue interactions. Binding of GRL0617 induces closure of the flexible blocking loop BL2 (Gly266 - Gly271) and narrows the substrate binding cleft. The residues Tyr268 and Gln269 in the BL2 loop shift towards the bound GRL0617.
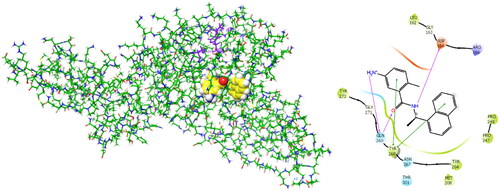
Table 3. Determined IC50 and EC50 and cytotoxicity of selected reference inhibitors of cysteine proteases.
Table 4. Determined IC50 and EC50 values of bis(benzylidene)cyclohexanones T97 - T125 and reference inhibitors of proteasome-associated deubiquitinating enzymes RA-9 and RA-190.
Figure 3. Evaluation of antiviral efficacy with calculation of the effective inhibitory concentration EC50 for compounds: T98 (A1, B1), T120 (A2, B2), T121 (A3, B3), T122 (A4, B4) and T124 (A5, B5). Monitoring the trend of decreasing the SARS-CoV-2 virus titre (normalised to untreated control, presented as mean with SEM) with increasing compound concentration (A1-A5). Determination of half-maximal effective concentrations EC50 [M] (B1-B5) from the sigmoidal model with the corresponding regression coefficient R2 (the goodness of fit). Data (B1-B5) are presented as mean and error with SEM.
![Figure 3. Evaluation of antiviral efficacy with calculation of the effective inhibitory concentration EC50 for compounds: T98 (A1, B1), T120 (A2, B2), T121 (A3, B3), T122 (A4, B4) and T124 (A5, B5). Monitoring the trend of decreasing the SARS-CoV-2 virus titre (normalised to untreated control, presented as mean with SEM) with increasing compound concentration (A1-A5). Determination of half-maximal effective concentrations EC50 [M] (B1-B5) from the sigmoidal model with the corresponding regression coefficient R2 (the goodness of fit). Data (B1-B5) are presented as mean and error with SEM.](/cms/asset/f1d0a03d-c7fb-4929-aa28-7a4bd4300842/ienz_a_2301772_f0003_b.jpg)
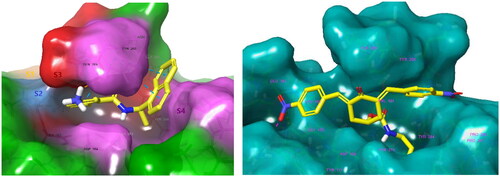
Figure 4. Left: Molecular surface of the binding site of GRL0617 that occupies the S4 - S3 pockets of the substrate binding cleft S4 - S3 - S2 - S1 of PLpro of SARS-CoV-2. The flexible BL2 loop G266-NYQC-G271 stabilises the bound ligand in its position by closing on the inhibitor (PDB entry 7CMD) Citation62. Right: Binding site of the model of noncovalent complex PLpro - T98. The partially transparent molecular surface of the enzyme illustrates the shape of the pockets of the substrate binding cleft. The amine tail is oriented downward and is not fully visible. Atom colouring scheme: H, white; C, yellow; N, blue; O, red. Only polar hydrogens are shown.