Figures & data
Figure 1. Chemical structures of Topo II inhibitors based on the carbazole scaffold (a–c), and symmetrically substituted carbazoles containing furan and thiophene (d).

Figure 2. The synthetic routes of the three carbazole derivatives (27a, 36a, 36b) (a). The structures of compounds 27a, 36a, and 36b, including atom numbering, are displayed to facilitate the interpretation of NMR spectra (b).

Figure 3. Inhibition of Topo IIα (a) and IIβ (b) mediated pBR322 relaxation. The experiment was carried out either with Topo IIα/IIβ in the presence of solvent (DMSO IIα/IIβ (+)) or with various concentrations of carbazole derivatives. ETP (100 µM), and ICRF-187 (100 µM) were used as references. (c) Unwinding assay. Determination of the ability of carbazole derivatives to intercalate into DNA in the presence of topoisomerase I (Top I). ETP and DOXO were used as negative and positive controls, respectively. The displayed gels have been cropped for clarity; full-length gels can be found in Supplementary Figures S1–S3.

Figure 4. Inhibition of Topo IIα/IIβ mediated kDNA decatenation by carbazole derivatives (a, b). DNA cleavage assay in the presence of 36a, 36b, and 27a, respectively (c, d). ETP (100 µM) and ICRF-187 (100 µM) were used as references. The displayed gels have been cropped for clarity; full-length gels can be found in Supplementary Figures S4–S7.

Table 1. In vitro growth inhibitory activity (IC50 ± SD, µM) of 27a, 36a, and 36bTable Footnotea.
Figure 5. Colony-forming ability of HCT-116 and A549 cells after treatment with 27a, 36a, and 36b. Representative images of the clonogenic assay for HCT-116 and A549 cell lines (a, b) and its quantification (c, d). Data represent the mean ± SD of three independent experiments.

Figure 6. Cytometric analysis of BrdU incorporation. A549, HCT-116, MCF-7, and U-2 OS cells were treated with 27a, 36a, and 36b for either 24 or 48 h. Representative histograms and statistical analyses after DNA staining (a). The results of the quantification analysis are presented in bar graphs. Error bars represent the mean ± SD of data obtained from three independent experiments (b).

Figure 7. Cell cycle profiles of A549, HCT-116, MCF-7, and U-2 OS cells after treatment with 36a, 36b, and 27a. Representative histograms from PI staining and their corresponding quantification are presented in the bar graphs. Error bars represent the mean ± SD of data obtained in three independent experiments.

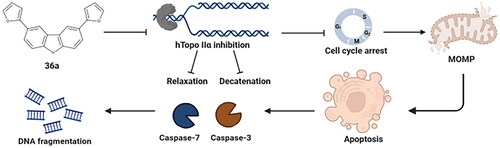
Figure 8. Analyses of proapoptotic activities of carbazole derivatives. Confocal imaging of A549 (a) and HCT-116 (b) cells after 24 h of treatment with 27a, 36a, and 36b. The cells were stained with Hoechst33342, scale bar = 10 µm. The arrows indicate cell shrinkage (white), fragmented nuclei (red), and apoptotic bodies (yellow). Quantification of flow cytometry analysis of A549 (c) and HCT-116 (d) cell lines (24 and 48 h) using Annexin V/7-AAD. Modulation of caspase 3/7 in A549 (e) and HCT-116 (f) cells after 24 and 48 h of treatment with 27a, 36a, and 36b. Analysis of changes in mitochondrial potential using JC-1 staining. Bar charts (g) with statistical quantification and confocal images of A549 and HCT-116 cells (h) acquired after 24 h of treatment with the compounds. Error bars represent the mean ± SD of data obtained from three independent experiments. ETP or FCCP served as references.

Table 2. Molecular formula, mass, and yield of the reagents and crude products 27a, 36a, and 36b.
Supplemental Material
Download MS Word (77 MB)Data availability statement
The datasets presented in the current study are available from the corresponding author upon reasonable request.