Figures & data
Figure 1. FDA-approved ALK and HDAC inhibitors and reported 2,4-pyrimidinediamine derivative ALK/HDAC dual inhibitor.

Figure 2. Design of benzimidazole dual ALK/HDAC inhibitors. The dashed boxes in the lead compounds indicated the preserved pharmacophore structure.

Scheme 1. Synthesis of compounds 3a, 3b, 3d, 3e. Reagents and conditions: (a) NH2XCO2Me or NH2XCO2Et, DIPEA, ACN, 80 °C, 12 h, 89–98%; (b) (i) NaOH, H2O, MeOH, rt, 12 h; iPrNH2, EDCI, HOBt, TEA, DMF, rt, 20 h, 88%; (c) ethyl acrylate, Herrmann’s palladacycle, [(tBu)3PH]BF4, Cy2NMe, DMF, MW, 30 min, 58–69%; (d) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (e) (i) benzoyl isothiocyanate, THF, 0 °C, 15 min; (ii) EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 36–75% (for step d and e); (f) (i) CNBr, EtOH, 25 °C, 16 h; (ii) RCOCl, TEA, DCM, 0 °C to rt, 2.5 h, 47% (for step d and f); (g) (i) LiOH, H2O, MeOH, rt, 20 h; (ii) NH2OTHP, EDCI, HOBt, TEA, DMF, rt, 20 h; (h) TFA, MeOH, rt, 6 h, 36–74% (for step g and h).
![Scheme 1. Synthesis of compounds 3a, 3b, 3d, 3e. Reagents and conditions: (a) NH2XCO2Me or NH2XCO2Et, DIPEA, ACN, 80 °C, 12 h, 89–98%; (b) (i) NaOH, H2O, MeOH, rt, 12 h; iPrNH2, EDCI, HOBt, TEA, DMF, rt, 20 h, 88%; (c) ethyl acrylate, Herrmann’s palladacycle, [(tBu)3PH]BF4, Cy2NMe, DMF, MW, 30 min, 58–69%; (d) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (e) (i) benzoyl isothiocyanate, THF, 0 °C, 15 min; (ii) EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 36–75% (for step d and e); (f) (i) CNBr, EtOH, 25 °C, 16 h; (ii) RCOCl, TEA, DCM, 0 °C to rt, 2.5 h, 47% (for step d and f); (g) (i) LiOH, H2O, MeOH, rt, 20 h; (ii) NH2OTHP, EDCI, HOBt, TEA, DMF, rt, 20 h; (h) TFA, MeOH, rt, 6 h, 36–74% (for step g and h).](/cms/asset/38b5bd12-ab0b-4eb6-8192-14f8e2de40cb/ienz_a_2318645_sch0001_b.jpg)
Scheme 2. Synthesis of compound 6. Reagents and conditions: (a) (i) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (ii) benzoyl isothiocyanate, THF, 0 °C, 15 min; EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 33%.

Scheme 3. Synthesis of compound 3c. Reagents and conditions: (a) methyl cis-1,4-aminocyclohexanecarboxylate hydrochloride, DIPEA, I, 80 °C, 12 h, 99%; (b) (i) NaOH, H2O, MeOH, rt, 12 h; (ii) iPrNH2, EDCI, HOBt, TEA, DMF, rt, 20 h, 41%; (c) ethyl acrylate, Herrmann’s palladacycle, [(tBu)3PH]BF4, Cy2NMe, DMF, MW, 30 min, 68%; (d) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (e) (i) benzoyl isothiocyanate, THF, 0 °C, 15 min; (ii) EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 63% (for step d and e); (f) (i) LiOH, H2O, MeOH, rt, 20 h; (ii) NH2OTHP, EDCI, HOBt, TEA, DMF, rt, 20 h; (g) TFA, MeOH, rt, 6 h, 75% (for step f and g).
![Scheme 3. Synthesis of compound 3c. Reagents and conditions: (a) methyl cis-1,4-aminocyclohexanecarboxylate hydrochloride, DIPEA, I, 80 °C, 12 h, 99%; (b) (i) NaOH, H2O, MeOH, rt, 12 h; (ii) iPrNH2, EDCI, HOBt, TEA, DMF, rt, 20 h, 41%; (c) ethyl acrylate, Herrmann’s palladacycle, [(tBu)3PH]BF4, Cy2NMe, DMF, MW, 30 min, 68%; (d) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (e) (i) benzoyl isothiocyanate, THF, 0 °C, 15 min; (ii) EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 63% (for step d and e); (f) (i) LiOH, H2O, MeOH, rt, 20 h; (ii) NH2OTHP, EDCI, HOBt, TEA, DMF, rt, 20 h; (g) TFA, MeOH, rt, 6 h, 75% (for step f and g).](/cms/asset/76c9c166-cf27-4932-9c01-bf8e1ba994f5/ienz_a_2318645_sch0003_b.jpg)
Scheme 4. Synthesis of compound 3f. Reagents and conditions: (a) NaN3, CuI, N,N-dimethylethylenediamine, NaCO3, DMSO, 110 °C, 5 h; (b) monomethyl suberate, EDCI, HOBt, TEA, DMF, rt, 20 h, 58% (for step a and b); (c) (i) Fe, NH4Cl, H2O, EtOH, 80 °C, 6 h; (ii) benzoyl isothiocyanate, THF, 0 °C, 15 min; (iii) EDCI, DIPEA, 60 °C, 2 h, then rt, 16 h, 65%; (d) (i) LiOH, H2O, MeOH, rt, 20 h; (ii) NH2OTHP, EDCI, HOBt, TEA, DMF, rt, 20 h; (e) TFA, DCM, 0 °C to rt, 12 h, 71% (for step d and e).

Table 1. Structure-activity relationships for test compounds (3a–3f and 6).
Table 2. Antiproliferative activity of compound 3b and 3c in cancer cell lines.
Figure 3. In vitro enzymatic activity of 3b against ALK mutants (IC50 in nM). No inhibitor control as 100% enzyme activity. The assay was tested in duplicated, and the results were indicated separately as black squares and hollow inverted triangles.

Table 3. In Vitro Enzymatic Activity of 3b Against Various Receptor Tyrosine Kinases (IC50 in μM).Table Footnotea
Table 4. In Vitro enzymatic activity of 3b against various ALK Mutants (IC50 in nM).Table Footnotea
Figure 4. Effect of test compounds on H2228 cells. Human NSCLC H2228 cell line was treated with 3b (2 µM), 3c (4 µM), or positive controls, SAHA as pan-HDAC inhibitor, pracinostat as class I and IIa HDAC inhibitor, and crizotinib as ALK inhibitor, with DMSO (as negative control) for 6 h. (A) Western blots of phosphorylated tyrosine residue 1640 on ALK as well as acetylated (Ac) α-tubulin and histone H3 with their corresponding total proteins for normalisation. GAPDH was used as a loading control. (B) Quantification of western blotting results using Image J. Error bars indicate ± S.D.E. *p-Value ≤ 0.05, **p-value ≤ 0.01, ***p-value ≤ 0.001.

Figure 5. The predicted binding mode for 3b in the binding site of ALKwt (PDB ID: 4MKC). (A) The yellow lines indicated hydrogen bonds and the estimated lengths of bonds was shown in purple number. (B) The purple arrows indicated hydrogen bonds.

Figure 6. The predicted binding mode for 3b in the binding site of HDAC6 (PDB ID: 5EDU). (A) The yellow lines indicated hydrogen bonds and the cyan lines indicated pi-pi interactions. (B) The purple arrows indicated hydrogen bonds, the green arrows indicated π–π interactions, and the gray line indicated metal interaction.

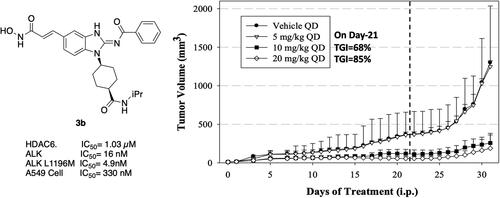
Figure 7. Tumour volume and body weight following treatment of human cancer A549 xenografts in mice with 3b. Indicated doses were administered daily up to day 21 (dashed line). Data are mean ± SEM.

Table 5. Inhibition of CYP450 Enzymes and hERG by 3b (IC50 in μM).Table Footnotea