Figures & data
Figure 1. Schematic representation of the experimental procedure. Lentiviral vectors carrying reprogramming factors were introduced into the hESM01 cell line, and the stable clones were selected for further analysis (zero transgene expression, genome stability, in vitro and in vivo pluripotency). The resulting hESM01-OSKMN-DOX-n5 cell line was differentiated into 3 types of somatic cells. Magnetically separated cells were reprogrammed by DOX induction and iPSC clones generated from each cell type were chosen for genome-wide analysis of reprogramming traces, somatic memory, and iPSC specific markers using transcription and DNA methylation data.
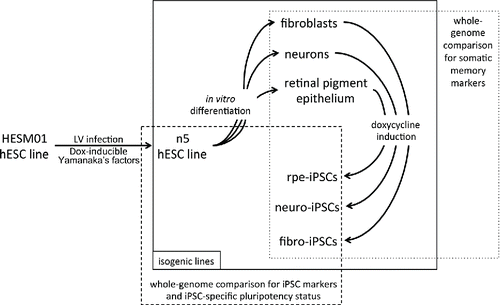
Figure 2. Characterization of n5 somatic derivatives. (A) Morphology (phase contrast microscopy) and immunocytochemistry of fibroblast-like cells (F), neurons (N), and retinal pigment epithelial cells (R). TUBb3, β-tubulin (III); CHAT, choline acetyltransferase; CK, cytokeratin; RPE65, retinal pigment epithelium-specific 65 kDa protein. Signals corresponding to the respective markers are shown in red, with blue indicating nuclei stained with DAPI. Scale bar, 100 mM. (B) FACS analysis of TRA-1-60 expression in differentiated cells. Dotted lines represent isotype controls and antibody staining is shown in green. (C) FACS analysis of the purity of magnetically separated F, N, and R cells for corresponding somatic markers.
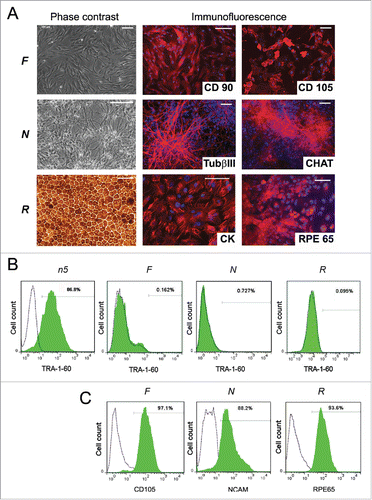
Figure 3. Characterization of isogenic iPSC lines reprogrammed from fibroblasts, neurons and RPE cells. (A) colony morphology of iF, iN, and iR cells; (B) immunocytochemical assay of iF, iN, and iR cells; for pluripotent markers (red and green indicate respective markers, blue indicates DAPI); (C) representative images of embryoid bodies and immunocytochemistry of in vitro iPSC-derived differentiated cell (red indicates markers, blue indicates DAPI) iF7 clones are shown. Scale bar, 100 mM; (D) Teratoma sections derived from the iF7 cell line, hematoxylin-eosin staining.
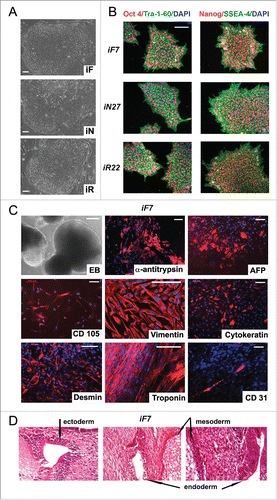
Figure 4. Genome-wide analysis of gene transcription and DNA methylation in the isogenic pluripotent and somatic cell lines. (A, B) Heatmaps based on Spearman correlations between all cell lines; the color scale ranges from 0.75 to 1. (A) Transcriptome and (B) methylome data (P < 0.01, FDR < 0.05).
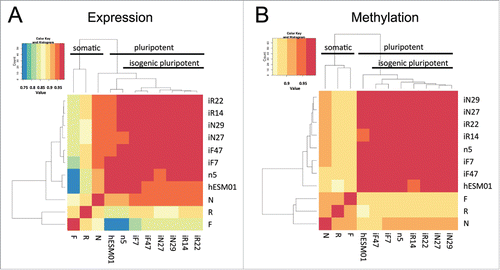
Figure 5. Reprogramming process, somatic memory and stochastic event input analysis in an isogenic system of human PSCs. (A) Graphical representation of the approach to analyze gene expression and CpG methylation during differentiation and reprogramming (“=” – same level of expression or methylation between indicated groups, “som” – somatic cell lines). (B) Input of genes (bold) and CpGs (italic) that drifted from specified groups (same colors as in A) into the “clone-specific” (red background) and “somatic memory” (blue background) groups during iPSC passaging. The number of passages is shown on top. (C) Clone-specific genes of different isogenic iPSC lines. (D) The methylation of 34 clone-specific CpGs is a specific reprogramming signature for our isogenic cell lines. The heatmap on the left shows that the methylation level of 34 CpG sites distinguishes iPSCs from ESCs and somatic cell lines in our isogenic system. However, the same CpGs do not discriminate ESCs and iPSCs in the independent GSE31848 data set; heatmap on the right. (E) Somatic memory group genes were identified for each iPSC type separately; however, most of them are shared by different iPSCs.
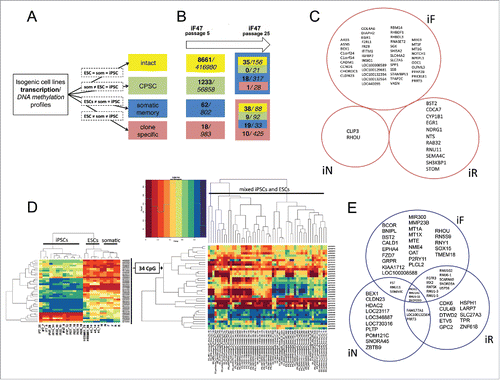
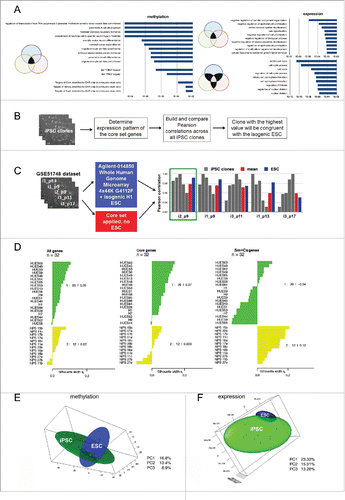
Figure 6. The isogenic system provides a core genetic signature for reprogrammed cells. (A) Top 10 Gene Ontology Biological Processes and Transcription Factor Targets shared by at least 2 different iPSC lines; the corresponding p-values are shown in bars. (B) Suggested approach to select an iPSC clone identical to its isogenic ESCs using the core gene set expression profile. (C) Examination of the best clone selection approach using the GSE51748 dataset. Pearson correlations between a particular iPSC clone and other clones calculated on the basis of the core set gene expression levels are shown in gray; the mean correlation between them is shown in red; Pearson correlations between particular iPSC clones and isogenic ESC lines calculated on the basis of all transcriptome data are shown in blue. The green rectangle indicates the best clone that has the highest mean correlation with others using both approaches. (D) Silhouette values (Sv) indicate the similarity of 32 ESCs (green cluster) and iPSCs (yellow cluster) lines from GSE25970 data set. On the right side, the Svs for each cluster are shown. Sv values were calculated using the expression pattern of all genes in the GSE25970 dataset (“All genes”): expression level only of the core set of genes; (“Core set”): expression level of the genes belonging to somatic memory and clone specific groups (“Sm+Cs genes”). (E, F) PCA analysis of isogenic PSC lines established in our study based on (E) methylation and (F) expression data. In both cases, 3 PCs were used and 3-D maps were built. The green sphere shows the location of the iPSC cluster, while the blue sphere shows the ESC cluster.