Figures & data
Figure 1. Network pharmacology and bioinformatics analysis to screen Panax notoginseng-related target genes for COPD regulation.
Note: (A) Venn diagram showing the intersection of Panax notoginseng’s related targets and COPD disease-related genes. (B) Interaction network diagram of proteins encoded by the 8 candidate target genes. (C) Bar chart showing the degree ranking of the 8 candidate target genes. (D) Importance ranking of the 8 candidate target genes in COPD as shown by the Phenolyzer database.
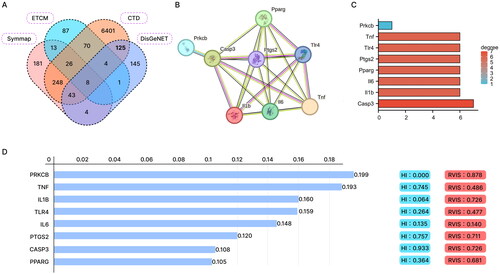
Figure 2. GO and KEGG enrichment analysis of candidate target genes.
Note: (A-C) GO function analysis shows the enrichment of the 8 candidate target genes in BP, MF, and CC. (D) KEGG pathway enrichment analysis of the 8 candidate target genes.
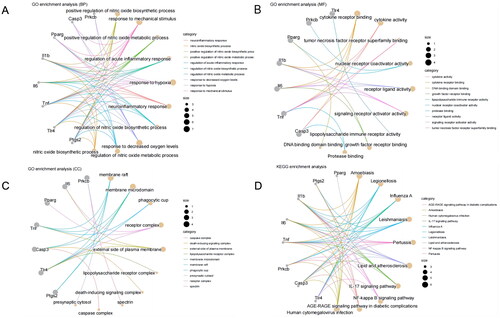
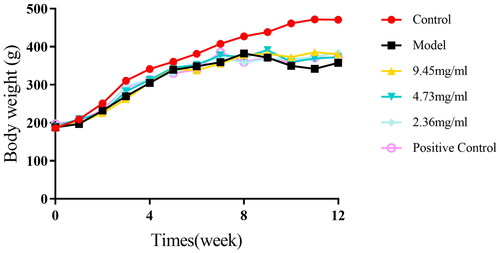
Figure 4. HE staining and lung function measurements were obtained for the lung tissue of each group of rats.
Note: A: HE staining results (10 × 10) of the lung tissue in each group of rats; B: Lung function (FEV0.2/FVC%, PEF) measurements in each group of rats; compared to the Control group, *P < 0.01, P < 0.05; compared to the Model group, ##p < 0.01, #p < 0.05.
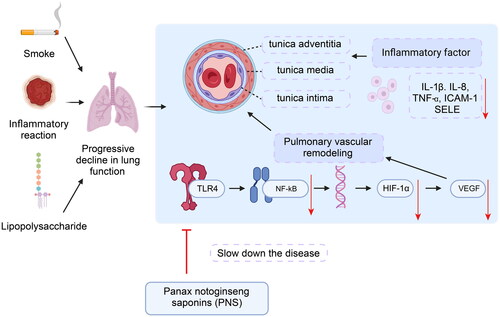
Figure 5. PNS’s effect on pulmonary vascular remodeling induced by COPD in rats.
Note: (A) Victoria + VG staining results of lung tissues from rats in each group (10 × 10). (B) Proportion of elastic fibers in lung tissues from rats in each group. (C) Proportion of collagen fibers in lung tissues from rats in each group. Compared to Control, **p <0.01, *p <0.05; compared to Model, ##p <0.01, #p <0.05.
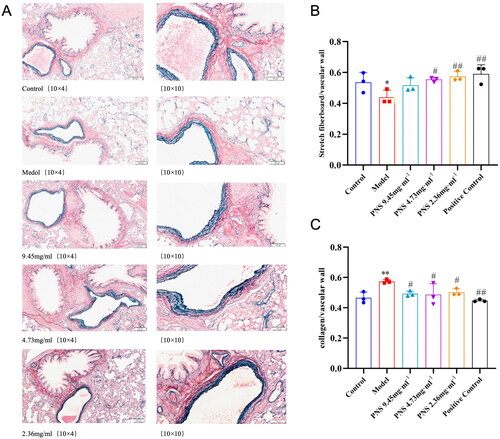
Figure 6. PNS affects the expression of related inflammatory factors and adhesion molecules in COPD rats.
Note: (A–C) ELISA detection of TNF-α, IL-8, and IL-1β levels in the serum of rats from each group. (D–E) ELISA detection of ICAM-1 and SELE levels in the serum of rats from each group. (F–G) RT-qPCR detection of ICAM-1 and SELE mRNA expression levels in lung tissues from rats in each group. Compared to Control, **p <0.01, *p <0.05; compared to Model, ##p <0.01, #p <0.05.
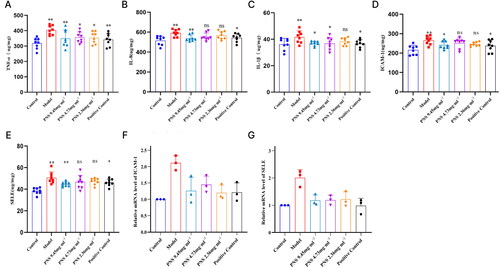
Figure 7. PNS’s regulation of TLR4/NF-κB/HIF-1α/VEGF pathway factor expression in COPD rats.
Note: (A) ELISA detection of VEGF concentration in the serum of rats from each group.
(B-D) RT-qPCR detection of TLR4, NF-κB, HIF-1α, and VEGF mRNA expression levels in lung tissues from rats in each group. Compared to Control, **p <0.01, *p <0.05; compared to Model, ##p <0.01, #p <0.05.
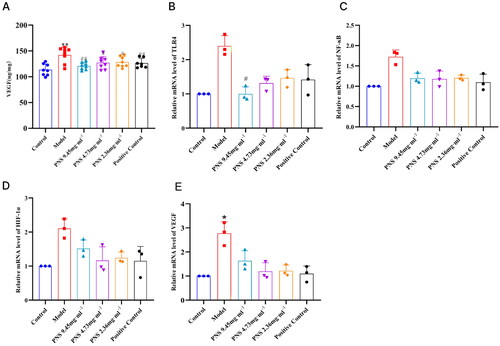
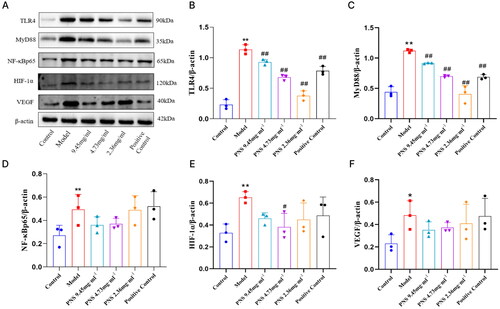
Figure 8. Changes in TLR4/NF-κB/HIF-1α/VEGF protein expression in lung tissues from rats in each group.
Note: (A) Western blot detection of TLR4, MyD88, NF-κB, HIF-1α, and VEGF protein expression in lung tissues from rats in each group. (B-F) Protein expression charts of TLR4, MyD88, NF-κB, HIF-1α, and VEGF in lung tissues from rats in each group. Compared to Control, **p <0.01, *p <0.05; compared to Model, ##p <0.01, #p <0.05.
Supplemental Material
Download Zip (6 MB)Data availability statement
The data underlying this article will be shared on reasonable request to the corresponding author.