Figures & data
Figure 1 Overexpressed Cx43-HKKSL is retained in the Golgi apparatus. HeLa cells stably transfected with Cx43-HKKSL at either low (a) or high (c) levels show differences in the pattern of intracellular localization by immunofluorescence microscopy. At low levels of expression, the HKKSL tag effectively functions to retain Cx43-HKKSL in the ER (a, arrowheads) and any Cx43-HKKSL which escapes into the Golgi apparatus is retrieved by the di-lysine tag (b). However, when overexpressed, Cx43-HKKSL remains outside the ER and accumulates in the perinuclear region of the cell (c, arrows) where it oligomerizes and forms large intracellular complexes that are resistant to retrieval (d) (Das Sarma et al. Citation2005). Alternatively, high levels of Cx43-HKKSL may force early oligomerization, which, in turn, promotes efflux into the Golgi apparatus and subsequent complex formation. Bar = 10 microns.
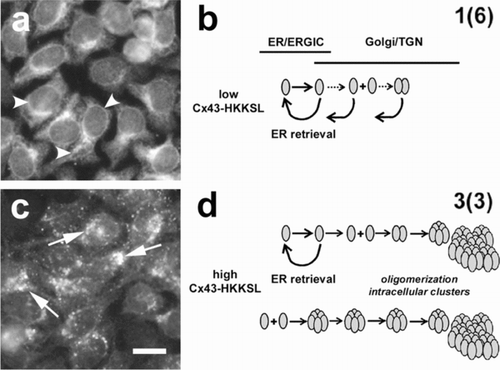
TABLE 1 Summary of cDNA screening
Figure 2 4-Phenylbutyrate restores ER localization of Cx43-HKKSL in 3(3) cells. HeLa cell transfectants stably expressing low (a, b; 1(6)) or high levels (c, d; 3(3)) of Cx43-HKKSL were treated for 18 h with 5 mM 4-PBA (a, c) or 0.1 μ g/ml geldanamycin (b, d), then fixed, permeabilized, and immunostained for Cx43-HKKSL. Following 4-PBA treatment, Cx43-HKKSL expressed by 3(3) cells reverted a distribution similar to 1(6) cells, showing enhanced ER localization (arrowheads). In contrast, geldanamycin had little, if any, effect on either 1(6) or 3(3) cells. Arrows show perinuclear localization of Cx43-HKKSL in 3(3) cells (d). Herbimycin A (3 μ g/ml) also had no obvious effect on either cell type (not shown). Bar = 10 microns.
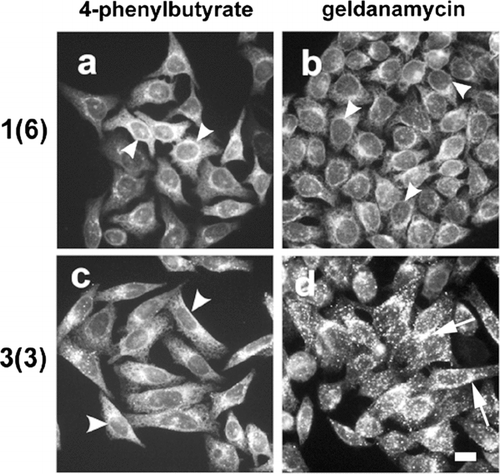
Figure 3 Cx43 did not compensate for Cx43-HKKSL overexpression. (a, b) Pools of cDNAs were screened for the presence of Cx43 cDNAs by transfection into HeLa cells, followed by immunofluorescence microscopy. Shown are two pools of clones from round 1 of screening, one of which contained Cx43 cDNA (a; pool 11), another that did not (b; pool 20). Arrowheads show Cx43 localized to gap junctions in (a). 3(3) cells transfected with myc-tagged Cx43 (Cx43-myc) did not show a change in Cx43-HKKSL localization (arrows), suggesting that Cx43 did not compensate for Cx43-HKKSL overexpression by 3(3) cells.
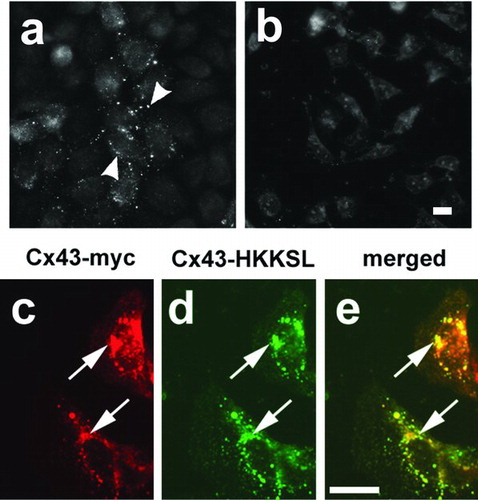
Figure 4 Assay of cDNA pools that compensate for Cx43-HKKSL overespression by 3(3) cells. Shown are examples of pools of clones which had high activity from round 1 (pool 20) containing ∼ 30,000 unique cDNA clones (a, b) and round 2 (pool 20-7) containing ∼ 1500 unique cDNA clones (c, d). (a and c) Fields of low activity where Cx43-HKKSL remained perinuclear (arrows); (b and d) areas of high activity where several 3(3) cells showed nuclear envelope localization of Cx43-HKKSL (arrowheads). Bar = 10 micron.
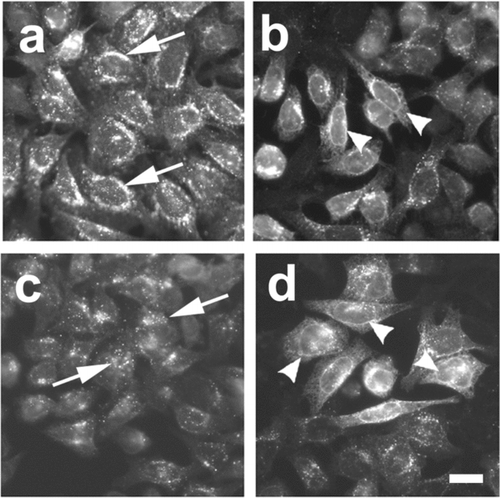
TABLE 2 Proteins identified in the screen
Figure 5 EGFP-tagged rab20 localizes predominantly to the perinuclear region of the cell. (a, b) Live HeLa cells transfected with EGFP-rab20 (a) or EGFP-rab20T44N (b) were analyzed by fluorescence microscopy. Most EGFP-rab20 localized to the perinuclear region of the cell (arrow), although there also was some nuclear envelope labeling (arrowhead), suggesting possible ER localization. EGFP-rab20T44N had a more uniform appearance, with low levels of apparent nuclear envelope labeling (arrowhead). Bar = 10 micron. (c) Immunoblot analysis of untransfected HeLa cells (un), cells transfected with EGFP-rab20 (wt) or EGFP-rab20T19N (T19N) using anti-EGFP for detection. Transfected cells show a specific band of near the predicted mass for the fusion proteins of ∼ 48 kDa.
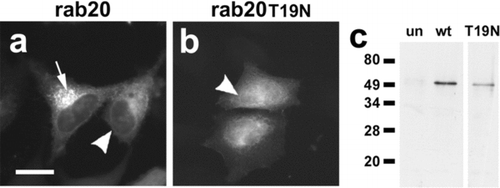
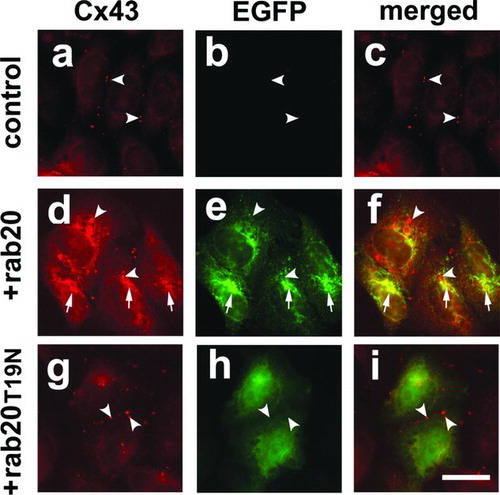
Figure 6 EGFP-rab20 inhibits Cx43 trafficking to the plasma membrane. Shown is immunofluorescence analysis of HeLa/Cx43 cells (a–c), HeLa/Cx43 cells transfected with EGFP-rab20 (d–f) or EGFP-rab20T19N (g–i). HeLa/Cx43 cells show Cx43 localized to gap junctions (arrowheads). Cells expressing EGFP-rab20 had partial co-localization with Cx43 in the perinuclear region of the cell (arrows), although there also were intracellular vesicles labeled for Cx43 that did contain EGFP-rab20 (arrowheads). By contrast, cells expressing EGFP-rab20T19N did not show this effect on Cx43, which assembled into gap junctions (arrowheads). Bar = 10 microns.