Figures & data
Figure 1 Cx45.6 degradation is primarily through proteasomal pathway. The lens primary cultures were treated with 20 μ g/ml of cycloheximide alone (A and B, control), cycloheximide plus lysosomal inhibitor, 100 μ M leupeptin (A), proteasomal inhibitor, 10 μ M clasto-lactacystin β-lactone (A), lysosomal inhibitor 10 mM NH4Cl (B), or proteasomal inhibitor, 0.5 μ M epoxomicin (B) for 0 h (lane 1), 4 h (lane 2), 10 h (lane 3), and 24 h (lane 4). The Cx45.6 protein bands from three separate Western blot analyses were quantified by densitometry (C). The data are presented as the mean ± s.e.m. (n = 3).
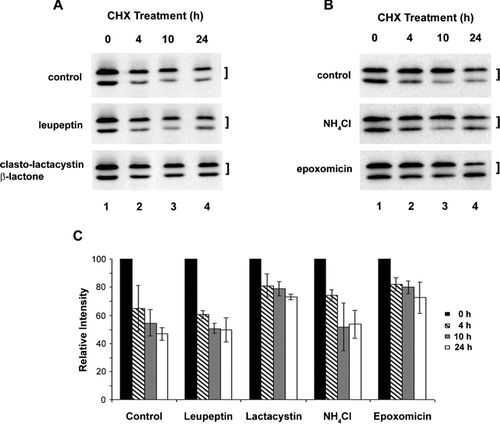
Figure 2 Co-expression of ubiquitin and connexin constructs in N2A cells. N2A cells were transiently transfected with wild type and mutant Cx45.6 constructs with or without ubiquitin. Cells were lysed 24 h after transfection. (A) Same amount of proteins from wild type Cx45.6 (lanes 1 and 3) and Cx45.6(S364A) (lanes 2 and 4) transfected cells co-expressing ubiquitin (lanes 3 and 4) or without ubiquitin (lanes 1 and 2) were immunoblotted by anti-flag antibody and anti-β actin antibody. The protein band intensity was measured by densitometry and the intensity ratio of Cx45.6 versus β-actin was calculated (right panel). (n = 3). (B) The transfected N2A cell lysates were immunoprecipitated by anti-Cx45.6 antibody. The immunoprecipitates from ubiquitin only (lane 1), wild type Cx45.6 (lanes 2 and 4) and Cx45.6(S364A) (lanes 3 and 5) transfected cells co-expressing ubiquitin (lanes 4 and 5) or without ubiquitin (lanes 1–3) were analyzed on 7.5% SDS-PAGE and immunoblotted with anti-myc and anti-flag antibodies, respectively. (n = 3).
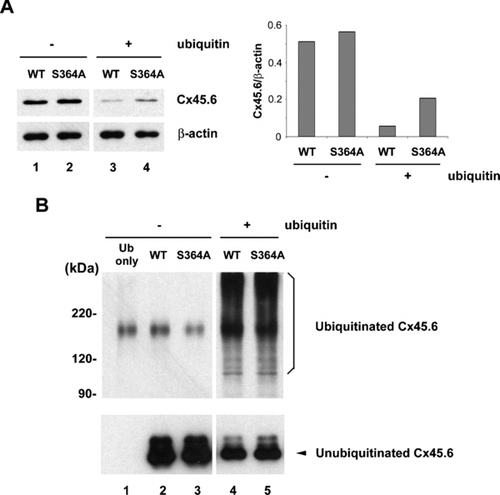
Figure 3 Constitutively phosphorylated mutant, Cx45.6(S364D), was resistant to the cleavage by caspase-3 as compared to wild type Cx45.6. Exogenous wild type (WT) Cx45.6 (lanes 1 and 2) and Cx45.6 single phosphorylation site mutants, Cx45.6(S364D) (lanes 3 and 4) were expressed in CEF cells through retroviral infection. Crude membranes isolated from those infected cultures were treated in the absence (lanes 1 and 3) and presence (lanes 2 and 4) of caspase-3 in vitro at 37°C for 6 h. Arrows indicate the cleaved fragments. The protein band intensity was measured by densitometry and the intensity ratio of truncated versus full-length Cx45.6 was calculated (lower panel). (n = 3).
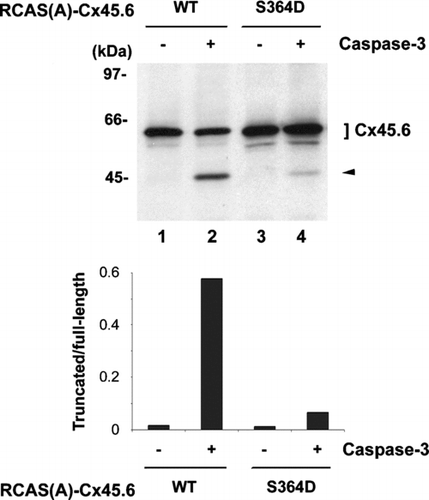
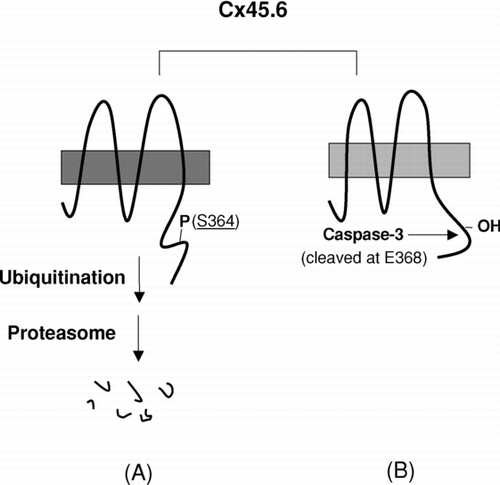
Figure 4 A model for the regulation of proteolysis of Cx45.6 in the lens by phosphorylation at Ser364. (A) The phosphorylation at Ser364 renders Cx45.6 unstable and undergoes the degradation mediated by proteasome. (B) If Ser364 is not phosphorylated, Cx45.6 becomes a substrate for caspase-3, which cleaves behind Glu368. The truncated Cx45.6 mainly accumulates in the mature fibers at the center core region of the lens (Yin et al. Citation2001).