Figures & data
Table 1. Primers used in experimental procedures.
Figure 1. (A). Analysis of AtHESP (At1G31500) promoter. EEs, CCA1 Binding Sites and E-boxes important for circadian regulation are marked. (B). Schematic representation of AtHESP (417 amino acids) showing the conserved nuclease domain (in blue) related to the Mg2+-dependent EEP superfamily, in comparison with the mouse NOC and human orthologHsCNOT6L. The latter possesses N-terminal leucine-rich repeats (LRRs) (shown in gray). The amino acid lengths are given for each polypeptide. The positions of the conserved catalytic amino acids are also indicated. (C). Partial AtHESP amino acid sequence alignment with characterized NOCs, focusing on conserved amino acid residues. HsNOC, MmNOC, RnNOC and XlNOC, orthologs of the EEP deadenylase family (HsCNOT6, HsCNOT6L and ScCCR4), and plant orthologs RcNGL1 and PtEEP, are included. The black line indicates the conserved PDILCLQEV domain for Mg2+/Mn2+ binding, which includes the catalytic residues E193 of MmNOC or E556 of ScCCR4 (both indicated by filled polygon). The filled square, the filled ellipse and the open star indicate catalytic residues D713, D780 and H818 of ScCCR4, respectively. Colors of residues are according to amino acid polarity. (D). Phylogenetic analysis of NOC, CCR4 and EEP protein sequences. The percentage of replicate trees in which the associated taxa clustered together is shown next to the branches. Scale bar, 0.1 substitutions per site. Hs, Homo sapiens; Ms, Mus musculus; Pt, Populus trichocarpa; Rc, Ricinus communis; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Xl, Xenopus laevis.
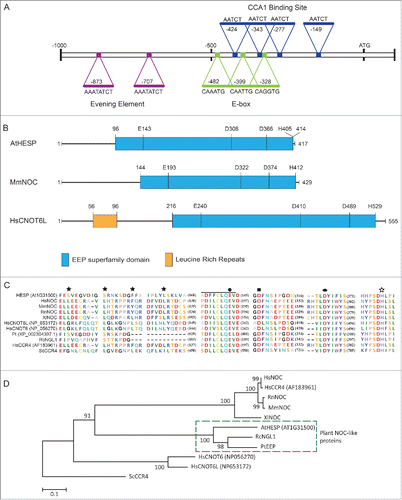
Figure 2. AtHESP is a Mg2+-dependent poly(A)-specific 3′ nuclease. (A). AtHESP degrades poly(A). Reaction rates of poly(A) degradation by AtHESP were measured using a colorimetric assay at the indicated substrate concentrations at 25°C (left panel) and 30°C (right panel), at 3 pH values (6.5, rhombus; 7.0, square; 7.5, circle). (B). AtHESP activity depends on Mg(II) ions. The rates in the absence (0mM), or excess (5mM) of Mg(II) were compared to those of 1.5 mM considered as the control. Mean values of 3 independent experiments. (C). Specificity for poly(A). The rates for poly(U), poly(C) and poly(G) were compared to those of poly(A). The reactions were performed with 600 mM of each polynucleotide at 25°C, pH 6.5, and 1.5 mM Mg2+. (D). AtHESP is a poly(A)-specific 3′ nuclease. L3A30 (filled arrow) or L3A30X49 (filled double arrow) RNAs were incubated with AtHESP (lanes 3-6) or PARN (lanes 7-10). The reactions were terminated at the indicated time points, denoted by numbers above the lanes (in minutes). The reacted RNA was purified and fractionated on 3% agarose gels stained with GelRed. Numbers on the left and the right indicate the positions of RNA length markers (nt). Filled arrow and double arrow indicate the positions of L3A30 (length 84 nt) and L3A30X49 (length 133 nt) RNA. Open arrowhead indicates the position of the reaction product.
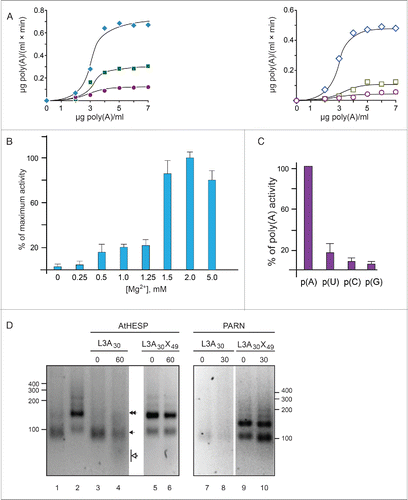
Figure 3. AtHESP is oligomeric. (A, B). Double reciprocal (A) and Hill (B) plots from data obtained from experiments as described in yielded slopes equal to 3 and 2.2, respectively. (C). SEC chromatogram. X axis, elution volume (ml); Y axis, A280 absorbance (arbitrary units; mAu). Asterisks indicate eluted proteins at certain molecular mass (*) 150 kDa, (**) 100 kDa. Arrows indicate the positions of mass markers: single arrow, glucogen phosphorylase (rabbit muscle) 97 kDa; double arrowhead, lysozyme 14.3 kDa. (D). Electrophoretic pattern of SEC fractions under denaturing conditions (10% SDS-PAGE). Numbers in red above the lanes correspond to actual numbering of ÄKTA fractions collected in . Numbers above lanes correspond to actual fraction numbering. Numbers on left indicate position of molecular mass markers (kDa) related to AtHESP size. IN: Input; M: molecular mass markers; E. Electrophoretic pattern of SEC fractions under non-denaturing conditions. Fractions were analyzed in a 7% polyacrylamide gel under non-denaturing conditions. Numbers in red above the lanes correspond to actual numbering of ÄKTA fractions collected in . Urease (Ur) and Bovine Serum Albumin (BA) were used as oligomeric structure and molecular mass markers; urease trimer (272 kDa) and hexamer (545 kDa), and BA monomer (66 kDa) and dimer (132 kDa). Arrows on the left of the gel indicate oligomeric structures, namely dimers, trimers and tetramers. Numbers on the right indicate positions of urease and BA oligomers and monomer.
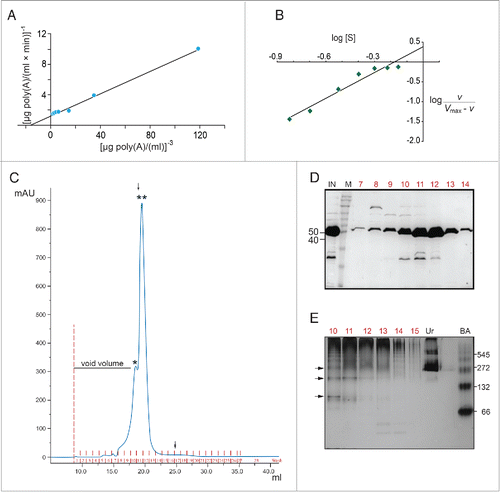
Figure 4. Diurnal and circadian time course of AtHESP, TOC1 and CCA1 expression in A. thaliana plants. A and B. Diurnal time course of expression levels in plants grown under 12 h/12 h light/dark (A) and 16 h/8 h light/dark (Β) photoperiod. Plants in (A) were transferred to constant light at day 7. Transcript levels were recorded every 4 hours. C and D. Diurnal expression of AtHESP in the triple toc1/lhy/cca1 (C) and toc1 mutant line (D) compared to wild-type plants. Plants were grown under 12 h/12 h light/dark photoperiod. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n = 3, each consisting of 15 plants). Asterisk (*) indicates statistically different mean values at each time point (t- test P ≤ 0.05). Three independent experimental trials yielded similar results.
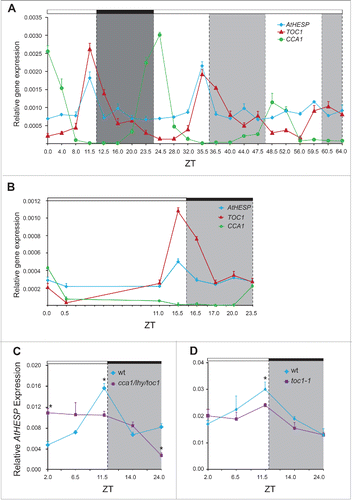
Figure 5. Diurnal gene expression and photosynthetic parameters in A. thaliana knockdown hesp plants. (A). Schematic representation of the insertion positions in the 3 T-DNA homozygous insertion mutants used in the present study. Colored boxes indicate exons and untranslated (UTR) regions. (B). Diurnal time course of AtHESP expression levels in 3 T-DNA homozygous insertion mutants hesp1-1, hesp1-2 and hesp1-3, compared to Col-1 wild type plants.Citation82 (C). Diurnal time course of AtHESP, TOC1 and CCA1 expression in hesp1-1 mutant plants. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n = 3, each consisting of 15 plants). (*) Indicates statistically different mean values at each time point relative to wild type (t- test P ≤ 0.05). Three independent experimental trials yielded similar results. D. Assessment of excitation energy flux at PSII in wild-type and hesp1-1 knock down mutant plants estimating the photochemical utilization (ΦPSII), the regulated heat dissipation (ΦNPQ), and non-regulated dissipation (ΦNO) parameters, the non-photochemical fluorescence quenching (NPQ), the excitation pressure (1-qp)representing the fraction of closed PSII reaction centers and the relative PSII electron transport rate (ETR). Plants were grown under 12 h/12 h light/dark photoperiod. Error bars represent standard error of means (n = 6). P values (**<0.01) indicate differences relative to wild type. The experiments were repeated twice.
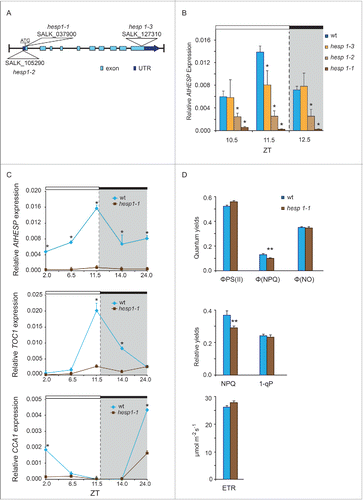
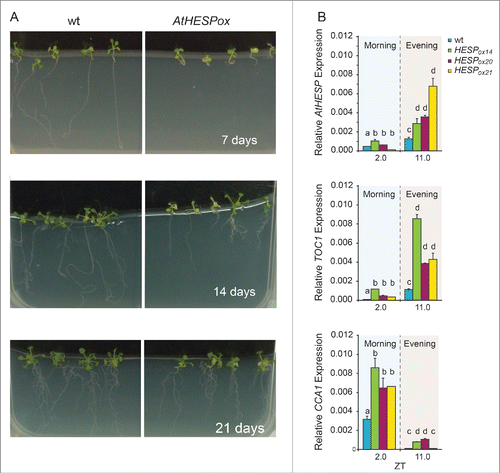
Figure 6. Diurnal time course of AtHESP, TOC1 and CCA1 expression in A. thaliana overexpression AtHESP-ox plants. (A). Growth and root length retardation phenotypes of AtHESP overexpressing plant lines (AtHESPox) (right panels) compared to wild type plants (left panels) grown for 7, 14 and 21 days in in vitro conditions. (B). AtHESP, TOC1 and CCA1 expression from 3 independent plant lines (AtHESPox14, AtHESPox20and AtHESPox21) with variably up-regulation of AtHESP compared to wild type (wt) plants at 2 time points is shown. Plants were grown under 12 h/12 h light/dark photoperiod. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n=3, each consisting of 15 plants). Statistical comparisons within plant lines were performed by Duncan tests (a <0.05). Indicator letters in common denote lack of significant difference. Three independent experimental trials yielded similar results.