Figures & data
Figure 1. Deamination of adenosine to inosine and ADAR family. (A) The schematic of adenosine-to-inosine deamination reaction catalysed by ADARs. (B) Domain structures of ADAR family genes in the vertebrate. There are three ADAR family members (ADAR1, ADAR2, and ADAR3), and ADAR1 contains two isoforms (p110 and p150). All the enzymes contain a conserved deaminase domain, shown in green, and the double-stranded RNA (dsRNA)-binding domain (dsRBD) that determines substrate specificity, shown in orange. ADAR1 p110 and p150 isoforms are different in their Z-DNA-binding domains, shown in blue. The only ADAR3 contains an arginine-rich domain, shown in purple
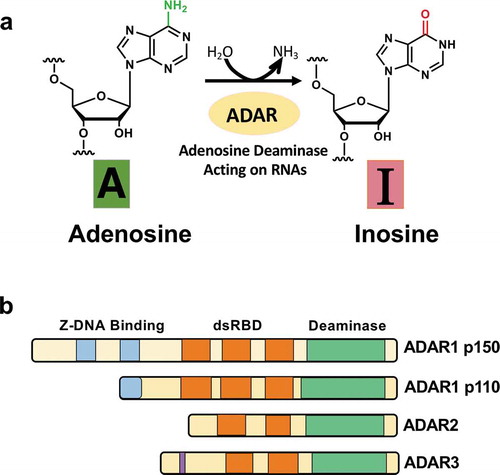
Figure 2. Functional roles of A-to-I RNA editing in the neurotransmitter receptor and the ion channel. (A) The A-to-I editing site (Q/R) in the secondary structure of the GluA2 pre-mRNA regions (top). The schematic of the GluA2 subunit structure and the functional difference in edited and unedited forms (middle and bottom, respectively). RNA editing alters the glutamate residue (Q) (middle left) at the 607th position to a positively charged arginine residue (R) (middle right), which can dramatically reduce calcium permeability (bottom). (B) The five editing sites (A-E) in the secondary structure of the 5HT2CR pre-mRNA regions where exon 5 base-pairs with the intron 5 (top). The schematic drawing of 5-HT2c receptor structure the functional difference in edited and unedited forms (middle and bottom, respectively). RNA editing of 5HT2C receptor converts amino acid isoleucine (I) at 156th position to valine (V), or methionine (M), asparagine (N) at 158th position to aspartic acid (D), serine (S), or glycine (G), and isoleucine (I) at 160th position to valine (V) (middle). Edited 5HT2CR isoforms exhibit reduced G protein coupling efficiency (bottom left) in comparison to the unedited isoform (bottom right). (C) The A-to-I editing site (I/V) in the secondary structure of Kv1.1 mRNAs (top). The schematic drawing of Kv1.1 channel structure and the functional difference in edited and unedited forms (middle and bottom, respectively) The transmembrane voltage sensor (S4), the K+ ion selectivity filter (S5–S6) are indicated in the middle and bottom figure. Editing of Kv1.1 changes the isoleucine (I) (middle) to valine (V) (bottom). which reduces the affinity for the binding of Kv β1.1 and enhances recovery from inactivation
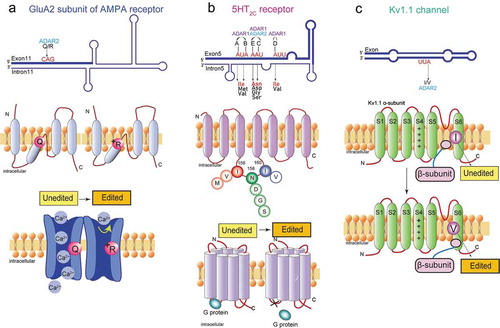
Table 1. Dysregulated A-to-I editing associated with neurological or neurodegenerative diseases
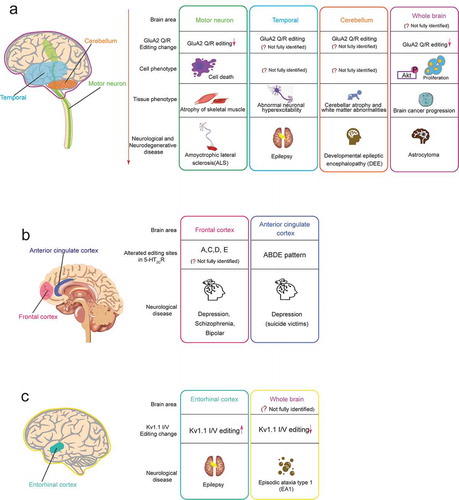
Figure 3. Neurological or neurodegenerative diseases associated with dysregulated A-to-I RNA editing in the neurotransmitter receptor and the ion channel. In each editing gene, the human brain areas affected by dysregulated RNA (including speculated areas) are shown in the left schematic drawing of the human brain, and associated diseases are shown in the right boxes. Each box includes the related brain area, alteration pattern of RNA editing, cell phenotype, tissue phenotype (only for GluA2 Q/R editing), and the associated disease. The colours of the outside line in each box corresponds to those of brain areas, respectively. (A) GluA2 Q/R editing (B) 5-HT2cR editing (C) Kv1.1 I/V editing