Figures & data
Figure 1. Hypoxia increases the number of acidic puncta and autophagosomes in nucleus pulposus (NP) cells. (A) Transmission electron microscopy images of primary NP cells cultured under normoxia (NX, 21% pO2) or hypoxia (HX, 1% pO2) for 24 h show multiple double-membrane enclosed autophagosomes (arrow); an increased number in hypoxia is observed. Scale bar: 500 nm. (B) Left panel: Immunofluorescence image of endogenous LC3 in NP cells shows increased LC3 puncta following 24 h in hypoxia. Scale bar: 50 μm. Middle panel: Acridine orange staining of NP cells shows cells cultured for 24 h in hypoxia have more acidic organelles (red signal). Scale bar: 35 μm. Right panel: LysoTracker® Red DND-99 staining of NP cells shows increased red staining after 24 h hypoxic culture. Scale bar: 50 μm. (C) Quantification of LysoTracker staining (red) as puncta area (px2)/cell using ImageJ software confirms increased acidic organelles under hypoxia. At least 427 cells per group were used for quantification analysis. (D) Western blot analysis of NP cells cultured under hypoxia for 8–72 h demonstrates accumulation of LC3-II protein at 8–48 h in hypoxia. (E) Densitometric analysis of multiple independent experiments as shown in the western blot confirms significant accumulation of LC3-II at 24 h hypoxia with an upward trend at both 8 and 48 h. Densitometric analysis of other autophagy-related proteins indicates that hypoxia does not influence levels of SQSTM1, BECN1, or the ATG12–ATG5 complex. Values shown are mean ± SE from at least 3 independent experiments. NS, nonsignificant; *, p < 0.05.
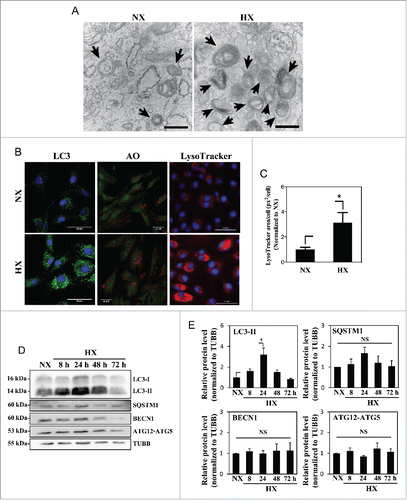
Figure 2. Hypoxia induces autophagy in NP cells without affecting its flux. (A) Western blot analysis of NP cells cultured in normoxia (NX) or hypoxia (1% and 5% pO2) with or without bafilomycin A1. Cells show accumulation of LC3-II under hypoxia, and further accumulation with bafilomycin A1 treatment confirms the presence of active flux under all culture conditions. (B) Densitometric analysis of multiple western blot experiments represented as both bar graphs and line graphs. Bar graph shows densitometry data normalized to respective untreated control group under each of the oxygen tensions. Inset line graph represents all densitometry data normalized to untreated normoxia control. Densitometric analysis confirms hypoxic accumulation of LC3-II. Both LC3-II and SQSTM1 further accumulated with bafilomycin A1, while no change was seen in the protein levels of BECN1 and the ATG12–ATG5 complex by hypoxia or bafilomycin A1. (C) NP cells transduced with retrovirus expressing a tandem EGFP-mCherry-LC3B construct cultured under either normoxia or hypoxia (HX) show autophagosomes in green-yellow and autolysosomes in red. Scale bar: 25 μm. The number of green-yellow puncta significantly increased under hypoxia, while the number of red-only puncta is more or less similar between normoxia and hypoxia. (D) Quantification of puncta area per cell using Colocalization Plugin of ImageJ software confirms significantly increased formation of autophagosomes with a relatively smaller change in autolysosomes. At least 48 cells per group were used for quantification analysis. Values shown are mean ± SE from at least 3 independent experiments. NS, nonsignificant; *, #, p < 0.05.
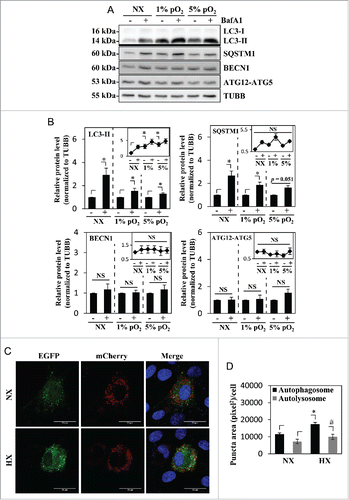
Figure 3. MTOR- and AMPK-dependent phosphorylation of ULK1 is unaffected by hypoxia. (A) Western blot analysis of NP cells cultured under normoxia (NX) or hypoxia (HX) for 8–72 h shows that the levels of p-ULK1 at Ser777 or Ser757 do not change with hypoxic culture. (B) Densitometric analysis of p-ULK1 Ser757 normalized to total ULK1 confirms MTOR-dependent ULK1 phosphorylation is unaltered under hypoxia until 24 h with very small decrease at 48 and 72 h. (C) p-ULK1 Ser777 level normalized to total ULK1 under hypoxia remains the same confirming AMPK phosphorylation of ULK1 is unaffected by hypoxia. Values shown are mean ± SE from at least 3 independent experiments. NS, nonsignificant; *, p < 0.05.
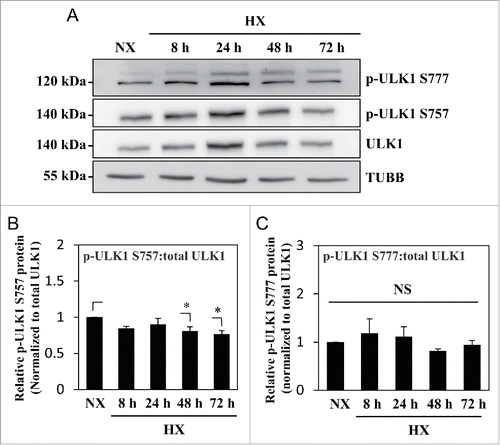
Figure 4. Autophagy in NP cells is unaffected by rapamycin treatment. (A) Immunofluorescence staining of endogenous LC3 in NP cells cultured under normoxia (NX) or hypoxia (HX), with or without rapamycin (500 nM) treatment shows hypoxia but not rapamycin increases LC3-positive puncta. Scale bar: 25 μm. (B) Acridine orange staining of NP cells shows increase in acidic puncta under hypoxia; no further increase is seen with rapamycin treatment. Scale bar: 35 μm. (C) Quantification of acridine orange signal (red) normalized to double-strand DNA signal (green) confirms there is no change in number of acidic compartments in NP cells treated with rapamycin. Bafilomycin A1 treatment, which inhibits acidification of lysosomes, was used as a negative control. (D) Western blot analysis of NP cells in either normoxia or hypoxia treated with an increasing dose of rapamycin (100, 250, 500 nM) shows no further increase of LC3-II with rapamycin treatment at all 3 doses. Similarly, levels of SQSTM1, BECN1, and ATG12–ATG5 are unaffected by rapamycin treatment. In addition, rapamycin does not change phosphorylation of ULK1 at Ser757, while completely abolishing phosphorylation of RPS6KB/p70S6K. (E) Densitometric analysis of multiple independent experiments confirms the western blot data. Values shown are mean ± SE from at least 3 independent experiments. NS, nonsignificant; Ctr, control; Rapa, rapamycin; BafA1, bafilomycin A1; *, p < 0.05.
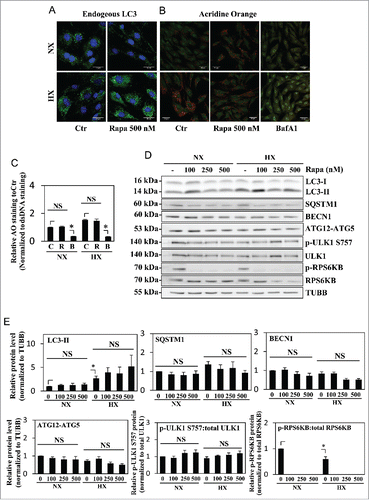
Figure 5. Autophagy in NP cells is regulated in an MTOR-independent fashion. (A) Western blot analysis of NP cells cultured under normoxia (NX) or hypoxia (HX) for 24 h, with or without Torin1 (200, 400 nM). LC3-II level is increased by hypoxia, but unaffected by Torin1 treatment. The p-ULK1 Ser757 level is significantly reduced with Torin1 treatment, confirming MTOR-dependent phosphorylation of ULK1 is successfully inhibited. The level of p-ULK1 Ser777 as well as total ULK1 is also reduced by Torin1, indicating Torin1 also affects MTOR function in protein synthesis. (B) Densitometric analysis of multiple independent western blot experiments confirms that inhibition of MTOR-dependent phosphorylation of ULK1 has no effect on LC3-II levels under both normoxia and hypoxia. (C) Immunofluorescence staining of endogenous LC3 demonstrates Torin1 has no effect on the number of LC3-positive autophagosomes in NP cells. Scale bar: 25 μm. (D) Western blot analysis of chondrocytes treated with either rapamycin (500 nM) or Torin1 (400 nM) for 6 h demonstrates increased LC3-II levels and concurrent decreased SQSTM1 levels. Both rapamycin and Torin1 blocked MTOR phosphorylation of ULK1 at Ser757. (E) Densitometric analysis of multiple independent western blot experiments confirms that modulation of MTOR activity alters the level of autophagy in chondrocytes. All the quantitative data are represented as mean ± SE from at least 3 independent experiments. NS, nonsignificant, Ctr/C, control; R, rapamycin; T, Torin1; *, p < 0.05.
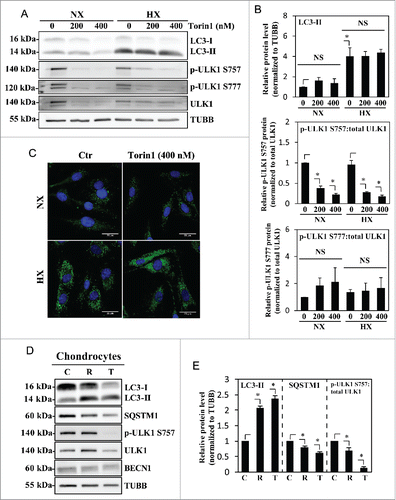
Figure 6. Hypoxic induction of BNIP3 does not correlate with the activation status of BECN1. (A) Western blot analysis of NP cells cultured under normoxia (NX) or hypoxia (HX) for 8–72 h shows that the BNIP3 level significantly increases by 24 h in hypoxia. BNIP3L level as well as p-BECN1 Ser93 did not change under hypoxia. (B) Densitometric analysis of western blot confirms hypoxic induction of BNIP3, peaking at 24 h. In contrast, the BNIP3L level remains relatively constant in hypoxia except for a small decrease at 72 h. Additionally, the level of p-BECN1 Ser93 is not affected under hypoxia, indicating a lack of correlation between BNIP3 and activation status of BECN1. All the quantitative data are represented as mean ± SE from at least 3 independent experiments. NS, nonsignificant; *, p < 0.05.
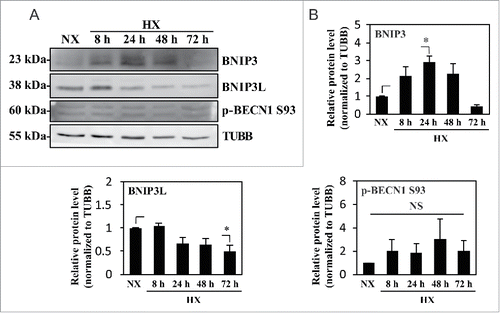
Figure 7. Hypoxic induction of autophagy in NP cells is independent of HIF1A activity. (A) YFP expression of NP cells transduced with lentivirus expressing ShCtr or ShHIF1A shows high ∼80–90% transduction efficiency. Scale bar: 100 μm. (B) Western blot analysis of NP cells transduced with ShCtr or ShHIF1A shows that HIF1A silencing does not affect LC3-II levels irrespective of oxemic tension. While BNIP3 levels also increase under hypoxia (HX) and decrease with HIF1A silencing, there is no appreciable change in both SQSTM1 and BNIP3L levels by hypoxia or HIF1A silencing. (C) Densitometric analysis of HIF1A, LC3-II, BNIP3, BNIP3L, and SQSTM1 western blot data shown in (B) confirms that HIF1A silencing has no effect on the level of LC3-II while decreasing the BNIP3 level. (D) Western blot analysis of NP cells transduced with either ShCtr or ShHIF1A, with or without bafilomycin A1 treatment under hypoxia shows that the LC3-II level and its further accumulation following bafilomycin A1 (Baf) treatment is unaltered by HIF1A silencing. (E) Densitometric analysis of western blot experiments (D) confirms a lack of effect of HIF1A silencing on autophagic flux. (F) Schematic diagram describing generation of NP-specific Hif1a conditional knockout mice. (G) Left panels: H&E staining of intervertebral disc from control and knockout mice at E15.5. Scale bar: 50 μm. Middle and right panels: Immunofluorescence staining of LC3 of NP tissue. LC3 staining demonstrates that lack of Hif1a has no effect on overall staining (middle panel) the number and distribution of LC3-positive puncta (arrowhead, right panel) in vivo, confirming the in vitro knockdown data. Scale bar: 20 μm. (H) Quantification of LC3 immunofluorescence staining of E15.5 disc tissue presented as number of puncta per μm3 shows similar number of LC3-positive autophagosomes between control and Hif1a mutant mice. n = for Hif1a mutant group, and n = 2 for control group. All the quantitative data are represented as mean ± SE from at least 3 independent experiments. NS, nonsignificant; *, p < 0.05.
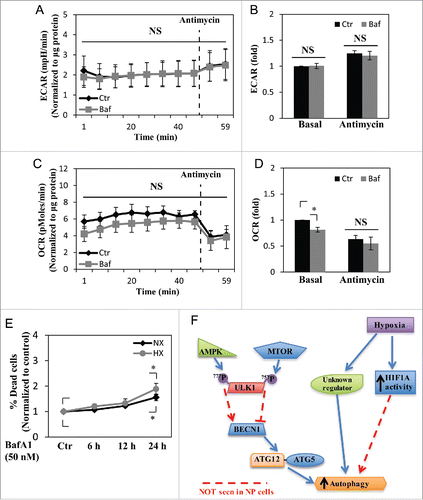
Figure 8. NP cell metabolism is unaffected by short-term inhibition of autophagy, but cell viability is reduced by long-term autophagy inhibition. (A and B) Extracellular acidification rate (ECAR) of NP cells treated with bafilomycin A1 (Baf) for 2–6 h under normoxia, measured by Seahorse XF Analyzer, shows no change in the ECAR with bafilomycin A1 treatment. (C and D) Oxygen consumption rate (OCR) of NP cells shows bafilomycin A1 treatment results in a slight reduction only in the basal OCR. (E) Cell viability test using Live/Dead assay demonstrates inhibition of autophagic degradation for 24 h results in decreased cell viability both in normoxia (NX) and hypoxia (HX). (F) Schematic diagram representing a model of noncanonical autophagy regulation in NP cells. Hypoxia induces a form of noncanonical autophagy in NP cells where formation of autophagosomes is affected more so than the autophagic flux. Moreover, autophagy in NP cells is largely independent of the MTOR-ULK1 axis. This noncanonical pathway also does not involve changes in the levels of autophagy-related proteins such as BECN1 and the ATG12–ATG5 complex. Interestingly, while hypoxia increases the activity of HIF1A, a known regulator of autophagy in some cell types, its activity has little or no effect on autophagy in NP cells. NS, nonsignificant.