Figures & data
Figure 1. Overview of the autophagic lysosome reformation pathway. (A) Autophagy commences with phagophore and autophagosome formation, which encapsulate cellular debris including damaged organelles and protein aggregates. Autophagosomes fuse with lysosomes forming autolysosomes where contents are degraded by lysosomal hydrolases. When starvation-induced autophagy is prolonged, the efflux of amino acids from autolysosomes initiates autophagic lysosome reformation (ALR). During ALR, the autolysosome membrane undergoes budding and extrusion to generate membranous tubules called “reformation tubules”. The scission of reformation tubules generates membrane fragments called proto-lysosomes, which mature into functional lysosomes. (B) Confocal laser scanning microscopy image of a reformation tubule formed at autolysosomes during ALR in an intact cell. As we described previously [Citation6], myoblasts were exposed to prolonged starvation-induced autophagy by culturing for 8 h in Earle’s balanced salt solution (EBSS). Cells were fixed and co-stained for LAMP1 to identify autolysosomes/lysosomes and DAPI to define nuclei. Each individual punctum present around the nucleus represents autolysosomes, and a single reformation tubule extends from an autolysosome, which is also shown at high magnification. Scale bar: 10 μm.
![Figure 1. Overview of the autophagic lysosome reformation pathway. (A) Autophagy commences with phagophore and autophagosome formation, which encapsulate cellular debris including damaged organelles and protein aggregates. Autophagosomes fuse with lysosomes forming autolysosomes where contents are degraded by lysosomal hydrolases. When starvation-induced autophagy is prolonged, the efflux of amino acids from autolysosomes initiates autophagic lysosome reformation (ALR). During ALR, the autolysosome membrane undergoes budding and extrusion to generate membranous tubules called “reformation tubules”. The scission of reformation tubules generates membrane fragments called proto-lysosomes, which mature into functional lysosomes. (B) Confocal laser scanning microscopy image of a reformation tubule formed at autolysosomes during ALR in an intact cell. As we described previously [Citation6], myoblasts were exposed to prolonged starvation-induced autophagy by culturing for 8 h in Earle’s balanced salt solution (EBSS). Cells were fixed and co-stained for LAMP1 to identify autolysosomes/lysosomes and DAPI to define nuclei. Each individual punctum present around the nucleus represents autolysosomes, and a single reformation tubule extends from an autolysosome, which is also shown at high magnification. Scale bar: 10 μm.](/cms/asset/58ff46f5-6270-4d63-be27-5cfb3408d8f2/kaup_a_2128019_f0001_oc.jpg)
Figure 2. Opposing roles of MTOR as a master regulator of both de novo lysosome biogenesis and autophagic lysosome reformation. Amino acid depletion during starvation results in MTOR inactivation, which induces autophagosome formation (1). MTOR inactivation also activates TFEB (and TFE3; not shown), which is dephosphorylated by PPP3/calcineurin and dissociates from YWHA/14-3-3 (2). TFEB (or TFE3) then translocate to the nucleus where they induce the transcription of genes required for de novo lysosome formation (3). Lysosomes fuse with autophagosomes to generate autolysosomes in which the contents are degraded and recycled (4). If autophagy activation is prolonged, the pervasive degradation of macromolecules within autolysosomes results in the release of amino acids into the cytosol via efflux transporters and also requires the function of the sugar transporter SPNS1/spin. The efflux of amino acids induces localized MTOR reactivation and initiates autophagic lysosome reformation (ALR) (5). Following initiation of ALR, reformation tubules form at autolysosomes and undergo scission to generate proto-lysosomes (6), that mature into functional lysosomes and can re-enter the autophagy pathway (7).
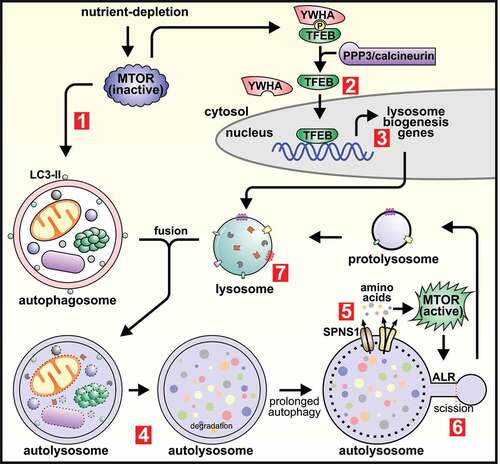
Table 1. Known and predicted phosphoinositide-regulatory enzyme and effector protein functions in ALR.
Figure 3. Membrane dynamics during autophagic lysosome reformation are under tight spatiotemporal control by phosphoinositides. (A) The ultrastructural membrane changes that take place at autolysosomes during ALR are driven by the localized generation of membrane-associated phosphoinositides, and are regulated by phosphatidylinositol-kinase and -phosphatase enzymes. PtdIns3P is generated from PtdIns by the class III phosphoinositide 3-kinase complex that includes PIK3C3/VPS34 (catalytic subunit), PIK3R4/VPS15 (regulatory subunit) and BECN1. In a successive pathway, the generation of PtdIns4P from PtdIns is controlled by the phosphatidylinositol 4-kinases PI4K2A or PI4KB/PI4K3B. In turn, PtdIns4P serves as a substrate for the synthesis of PtdIns(4,5)P2 by the PtdIns4P-5kinases, PIP5K1A or PIP5K1B. Interconversion between PtdIns(4,5)P2 and PtdIns4P on autolysosome membranes is important for ALR, whereby PtdIns(4,5)P2 is hydrolyzed by the inositol polyphosphate 5-phosphatase INPP5K to form PtdIns4P. (B) The generation of PtdIns(4,5)P2 at discrete foci on the autolysosome membrane initiates a succession of ultrastructural changes through the recruitment of specific effector proteins. This commences with the AP-2-clathrin complex which stimulates membrane budding, followed by KIF5B and WHAMM to extrude membrane buds into reformation tubules. KIF5B a motor protein, “pulls” membranes along microtubules, and WHAMM forces membranes into a tubular structure by stimulating localized actin polymerization. BLOC1S1 interacts with both KIF5B and WHAMM to co-ordinate this membrane extrusion stage. SACS is also important for reformation tubule formation possibly through its regulation of microtubule dynamics and/or binding to the AP-2-clathrin complex and KIF5B. DNM2 drives the scission of reformation tubules to form proto-lysosomes. MTOR activation of UVRAG (UV radiation resistance associated)-PIK3C3/VPS34 results in generation of PtdIns3P from PtdIns, which has also been implicated in the scission of reformation tubules to generate lysosomes. The hereditary spastic paraplegia protein complex AP-5-SPG11-ZFYVE26, is recruited to autolysosomes via ZFYVE26 binding to PtdIns3P and is required for ALR. ZFYVE26 also binds PI4KB/PI4K3B and has a role in regulating PtdIns4P at autolysosomes.
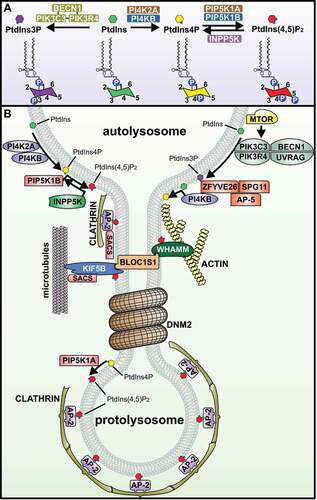
Table 2. Disorders showing inhibition of autophagic lysosome reformation.

