Figures & data
Figure 1. Characterization of proteins interacting with AtMed15a KIX domain. (A) Yeast two-hybrid screening was done using AtMed15a KIX as the bait. False positive clones were characterized by yeast two-hybrid analysis using vector alone (BD) as a control (left panel). Cultures of co-transformed yeast were spotted on SD Trp−/Leu− to monitor the growth and on SD Trp−/Leu−/His−/Ade− to score the interactions. Right panel shows clones of AtMed15a KIX-domain interacting protein (BD KIX). (B) Interaction of selected proteins with AtMed15a KIX domain (upper panel) and full-length AtMed15a (lower panel) was validated by BiFC analysis. AtMed15a KIX, AtMed15a and Y2H positive clones (MYB63, R3H and UKTF1) were cloned in compatible YFP BiFc vectors and bombarded on onion peel. YFP and DAPI (blue) fluorescence was observed by confocal microscopy. DAPI staining was used to identify nuclei in the cells.
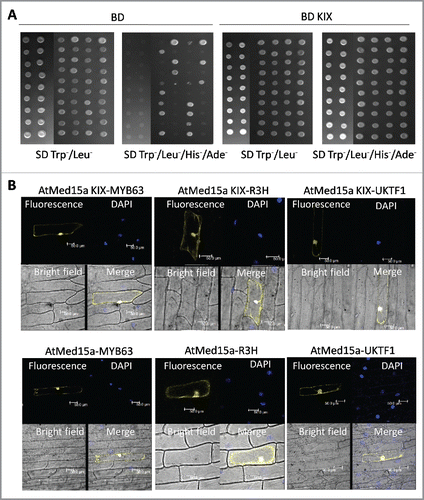
Figure 2. The interactome of AtMed15a KIX represents diverse proteins with different functions. (A) Graphical representation of the functional categories of proteins found to interact with AtMed15a KIX. Note that interactors with multiple functions can be present in different categories (Listed in Table S4). (B) AtMed15a was cloned in YFP vector, bombarded on onion peel and visualized using confocal microscopy.
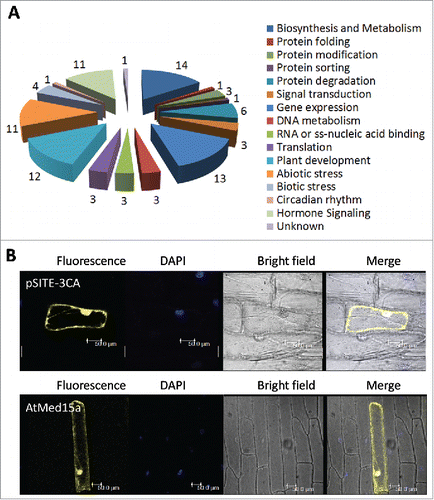
Figure 3. AtMed15a KIX interacting region of MYB63 harbors 9aa TADs. (A) Y2HGold co-transformed with derivatives of AD MYB63 clones and BD KIX or BD vector as control were spotted and grown on SD Trp−/Leu−/His−/Ade− with 10 mM 3-AT and 125 ng/ml Aureobasidin A agar media to score interactions. (B) 9aa TAD regions of AtMed15a KIX-interacting region of MYB63 are highlighted in red.
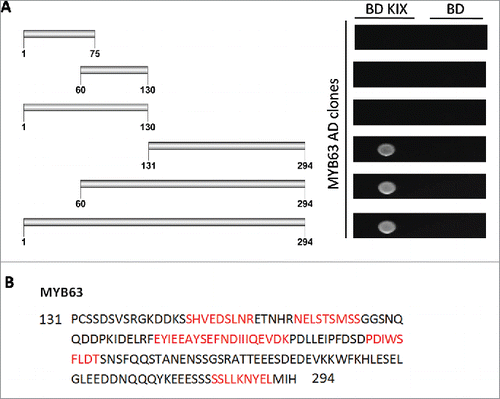
Figure 4. AtMed15a KIX interacting region of UKTF1 harbors 9aa TADs. (A) Y2HGold co-transformed with derivatives of AD UKTF1 clones and BD KIX or BD vector as control were spotted and grown on SD Trp−/Leu−/His−/Ade− with 10 mM 3-AT and 125 ng/ml Aureobasidin A agar media to score interactions. (B) 9aa TAD of AtMed15a KIX-interacting regions of UKTF1 are highlighted in red.
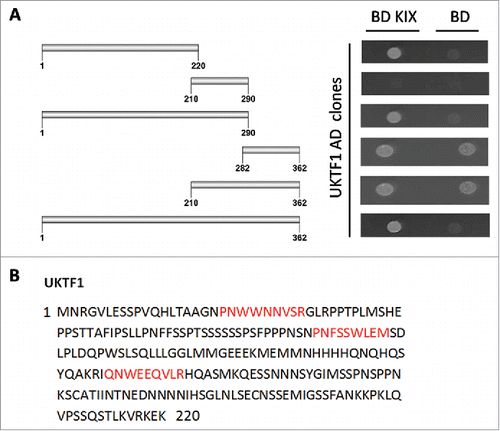
Figure 5. Transactivation domain of UKTF1. (A) Fragments, of AtMed15a KIX-interacting region of UKTF1 (U1 to U6), were cloned in pGBKT7 (BD) vector. (B) Y187 yeast cells were transformed with BD derivative clones (U1 to U6) of UKTF1 or BD vector control (V) and processed for β-Galactosidase assay. Graph showing β-gal units of untransformed Y187 (Y), Y187 transformed with BD vector (V) or BD clones of UKTF1 derivatives (U1-U6).
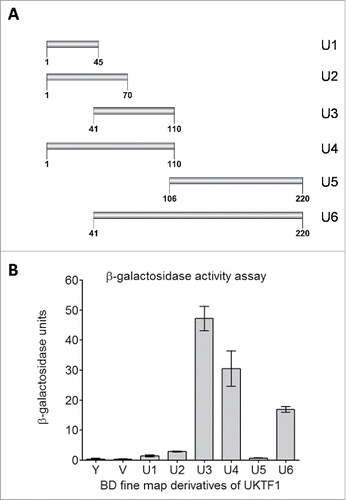
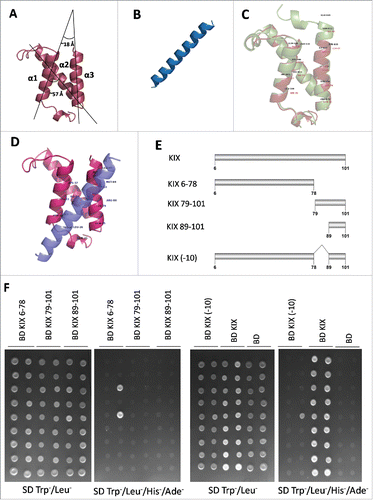
Figure 6. Third helix of AtMED15a KIX is required for protein-protein interactions. (A) Structure of AtMed15a KIX was predicted by homology modelling using mCBP KIX as template. (B) Based on the presence of binding region and 9aaTADs, MYB63 (D176 to D203) peptide was selected for docking studies. (C) Structures of mCBP KIX (green) and AtMed15a KIX (brown) were aligned using PyMOL, Labels show interacting residues. (D) Complex of AtMed15a KIX with MYB63 (D176 to D203), showing interacting residues of AtMed15a KIX. (E) Fragments of AtMed15a KIX were made based on the prediction of interacting residues in α3. (F) Interaction of Y2H positive clones with different fragments of AtMed15a KIX domain, as mentioned in (E), was checked. Cultures of co-transformed yeast were spotted on SD Trp−/Leu− to show proper growth, while on SD Trp−/Leu−/His−/Ade− to score the interactions.