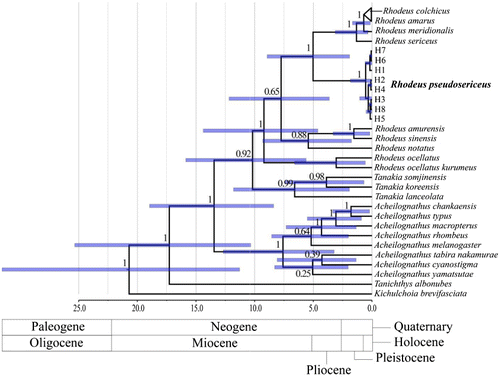
Figures & data
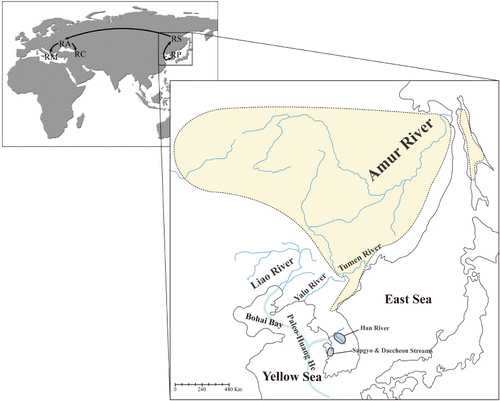
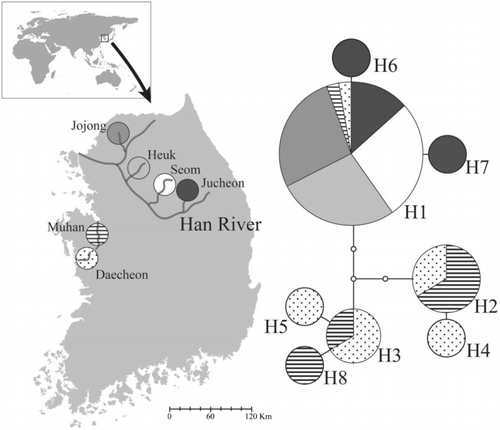
Table 1. List of the six R. pseudosericeus populations used in the present study.
Table 2. GenBank accession numbers and the references of the haplotypes used in the BEAST species-tree reconstruction based on fossil calibration at the stem of Acheilognathinae lineage and a substitution rate for the cyt b gene ().
Table 3. Summary of the AMOVA results that partition genetic variation among six R. pseudosericeus populations from the Korean Peninsula to two different geographic levels (among groups: the Han River vs. Muhan-Daechon).