Figures & data
Table 1. Antibody specifications.
Figure 1. Characterisation and quantification of platelet EVs. SSC-H over FSC-H colour dot plots: water and 200 beads/µl were recorded. Background noise (light blue) and bead gate (red) was set (a). Platelet EV suspensions without or with CaCl2 were recorded with a limit set on bead gate (b). Colour dot plots of platelet EV isotype control (blue) (c) and annexin A5/CD41a positive platelet EVs (green) (d). Size distribution of platelet EV suspensions measured by advanced nanoparticle tracking analysis (n = 10) (e).
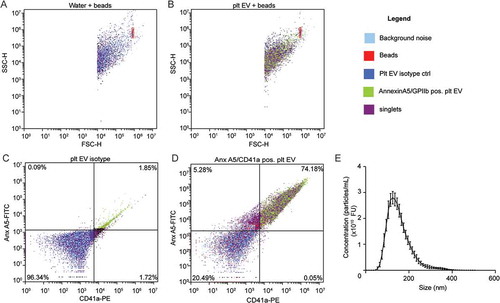
Figure 2. Cryo-transmission electron microscopy and bead coupled flow cytometry of platelet EVs. Representative micrographs from cryo-TEM of platelet EV suspensions. Scale bar 200 nm (a). Representative scatterplots and quantification of bead coupled flow cytometry for CD9 (b), CD63 (c) and CD81 (d) (n = 3).
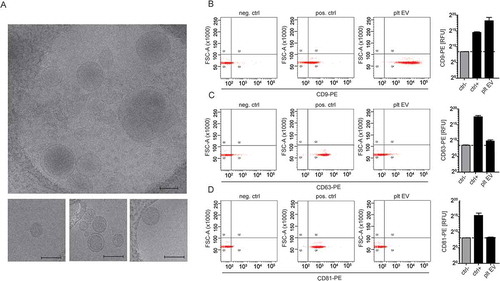
Figure 3. Platelet EVs interact with resting or TNFα stimulated SMC via platelet integrin αIIbβ3 or CX3CR1. CFSE-platelet EVs bound to SMC were measured by flow cytometry. Representative histograms of CFSE- platelet EVs bound to resting SMC in the presence of EDTA (filled histogram) or CaCl2 (line histogram) (a). CFSE-labelled platelet EVs (2 × 108/ml) were co-incubated with resting or TNFα-stimulated SMC in presence or absence of indicated inhibitors (eptifibatide, F1-Fk, P-selectin inhibitor) or blocking antibodies (CD40L) for 30 min at room temperature. Median fluorescence intensity of CFSE-platelet EV bound to resting (b) or TNFα-stimulated SMC (c) and platelet receptors implicated in platelet EV–SMC interaction. p-values were calculated by ANOVA with Sidak’s post-test (n = 3–7).
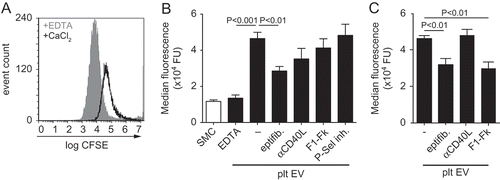
Figure 4. Platelet EVs change SMC morphology. Micrographs of SMC stained with antibodies against F-actin (a), αSMA (b) or calponin (c) after control, platelet EV-, PDGF- or CXCL4-stimulation for 96 h. Nuclei were stained with DAPI. Scale bars: 50 µm. Cell surface area (µm2) (d,e) and corrected total cell fluorescence (CTCF) (f) were quantified and displayed as bar graphs or box-and-whiskers plots, respectively. p-values were calculated by ANOVA with Tukey’s (d,e) or Kruskal–Wallis (f) post-tests (n = 3).
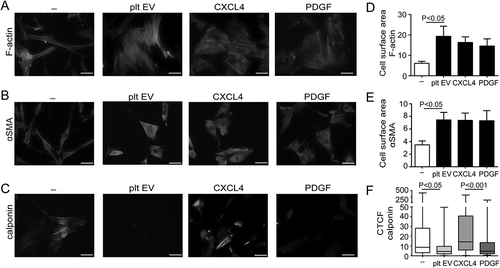
Figure 5. Platelet EVs induce migration and proliferation of SMC. Proliferation of SMC was measured following platelet EV–SMC co-incubation in absence and presence of indicated blocking antibodies; n = 3 (a). SMC migration was assessed in a Boyden chamber in the presence of medium, Glioma-EV, platelet EVs, SEC-platelet EVs, PDGF, CXCL4, CCL5 or CD40L; n = 4–12 (b) or blocking antibodies (PDGF), heparin or heparinase III; n = 3–8 (c). Dose-dependent SMC migration of CXCL4 reconstituted platelet EVs (d). P-values were calculated by ANOVA with Tukey’s post-test.
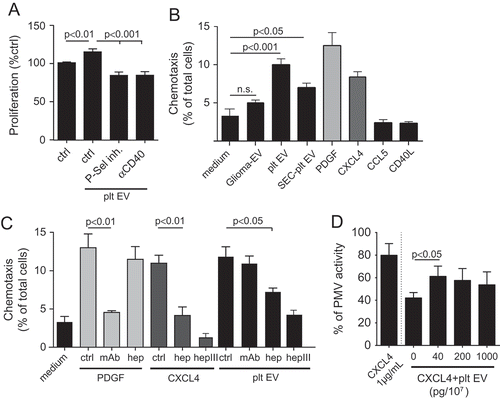
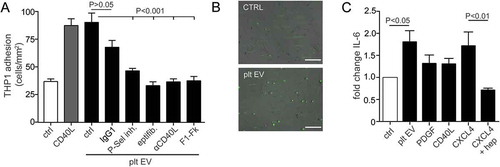
Figure 6. Platelet EVs increase monocytic cell adhesion and cytokine release. SMC were incubated without or with platelet EV or CD40L in the presence of indicated inhibitors (P-selectin inhibitor, eptifibatide, F1-Fk) or blocking antibody (α-CD40L). Syto13 fluorescently labelled THP-1 were perfused at 0.15 ml/min (3 dynes/cm2) and adherent cells were quantified in six different fields; n = 5–9 (a). Representative micrographs of THP-1 cells adherent to non- or platelet EV-stimulated SMC (b). SMC were incubated with indicated agonists for 24 h and IL-6 release was measured by ELISA; n = 6 (c). P-values were calculated by ANOVA with Tukey’s post-test. Scale bar: 100 µm.