Figures & data
Figure 1. Small RNA sequencing analysis in Fusarium oxysporum f. sp. cubense (Foc). (a) Classification and annotation of sRNAs. (b) sRNA length distribution. (c) and (d) Statistical analysis of initial bases in sRNA from XJZ2-culture (c) and XJZ2-36 hpi (d).
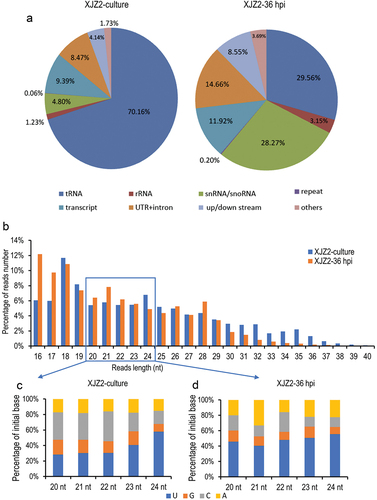
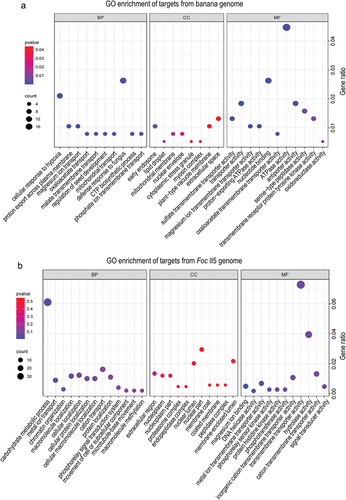
Figure 2. Identification of infection-induced milRNAs in Fusarium oxysporum f. sp. cubense. (a) Differential expression analysis of sRNAs of Foc in the pure culture stage and the initial infection stage. (b) Precursors’ secondary structure and sequences of infection-induced milRNAs in Foc. The mature milRNA sequences were marked in light blue. (c) Expression of the milRNAs in pure culture stage and initial infection stage by qRT-PCR. A Student’s t-test was used for significant analysis. **, p < 0.01. Error bars indicate S.D. (n = 3).
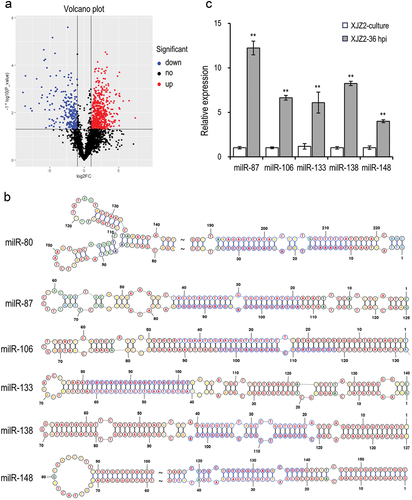
Figure 3. Expression of the milRNAs in different gene deletion mutants involved in the sRNA biogenesis pathway in Fusarium oxysporum f. sp. cubense. Transcriptional levels of the infection-induced milRNAs were detected by qRT-PCR. A Duncan’s multiple range test was used for significant analysis. Error bars indicate S. D. (n = 3). Different letters on the bars indicate significant differences at the level of α = 0.01.
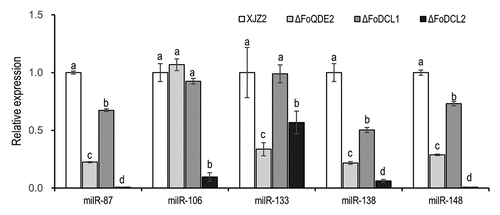
Figure 4. Verification of the milR106 deletion and complementation mutants by PCR and qRT-PCR. (a) Schematic diagram for deletion and complementation of milR106 in Foc. (b) PCR identification of the milR106 deletion mutants and complemented strains. (c) Transcriptional level of milR106 in different mutants of Foc was detected by qRT-PCR. To assess the statistical significance between the wild-type strain XJZ2 and the ΔmilR106-4 mutant, a Student’s t-test was used. ***, p < 0.001. Error bars indicate S.D. (n = 3).
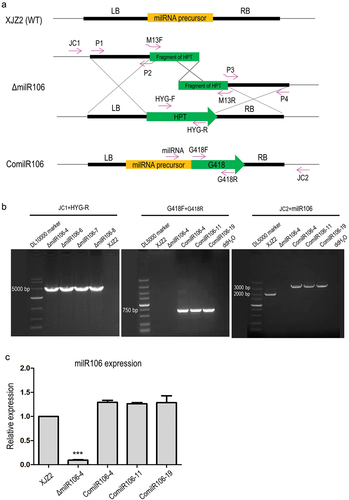
Figure 5. Characterization of milR106-deleted mutant and complemented strains in Fusarium oxysporum f. sp. cubense. (a) Colony morphology of the WT strain XJZ2, the ΔmilR106 mutant (ΔmilR106-4), and milR106 complemented strain ComilR106-19 on PDA plates. (b) Mycelial sensitivity to H2O2. Mycelial growth of the above strains was measured at MM and MM with 3 mmol/L H2O2. Bar=1 cm. (c) Percentage of mycelial growth inhibition by H2O2. A Student’s t-test was used for significant analysis. *, p < 0.05. Error bars indicate S.D. (n = 3).
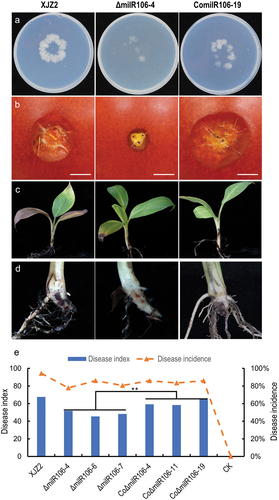
Figure 6. Assessment of invasive growth and Pathogenicity. (a) Cellophane penetration assay comparing the invasive growth of the WT strain XJZ2, ΔmilR106-4 mutant, and the complemented strain ComilR106-19. Conidia suspensions (100 μL per strain) with the same concentration of 1 × 105 spores/mL were put on cellophane-covered PDA plates and incubated at 28 °C for 4 days, then the cellophane sheets were removed, and samples were incubated for an additional 3 days and the colony size indicated mycelial growth were photographed. (b) Invasive growth on tomato fruits. The surfaces of tomato fruits were inoculated with discs of the different strains. The symptoms of necrosis on tomato fruits were recorded at 5 days post-inoculation. Bar =1 cm. (c) and (d) Symptoms of pathogenicity on the leaves (c) and corms (d) of the banana plantlets. (e) Disease index and disease incidence of banana plantlets after inoculation with the different strains. A Student’s t-test was used for significant analysis. **, p < 0.01.