Figures & data
Figure 1. CTA1 evokes near maximal cellular immune responses after a single vaccination: Groups of 6 BALB/c mice were immunized 1, 2 or 3 times with or without the CTA1 adjuvant on days 0, 14 and/or 28 (i.e., mice vaccinated twice were vaccinated on days 14 and 28 and mice vaccinated once were vaccinated on day 28). Mice were inoculated with 30 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 25 μg of empty plasmid as a no adjuvant control or 25 μg of pCTA1. ELISpot analysis was performed on day 48. Day 48 correlates to 20 d post the final immunization for mice vaccinated 2 or 3 times and 20 d post the single immunization for mice vaccinated a single time. (A) Splenocytes were assayed on day 48 by IFN-γ ELISpots with peptide pools encompassing SIVmac239gag or HIVBaLgp120. (B) Anti-gp120 reciprocal half maximal IgG titers were determined on day 48 sera by ELISA. The error bars represent the standard errors of the means. The P values (compared to the matching no adjuvant group) were calculated with a Students T Test using SigmaPlot v11 software. NS stands for not significantly different from the unadjuvanted control. The results shown are from a single experiment performed.
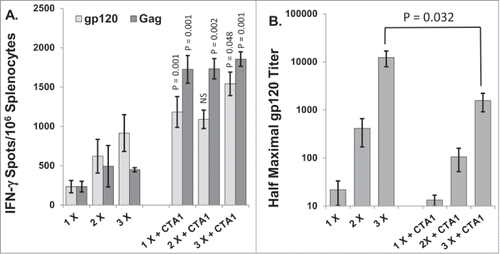
Figure 2. CTA1 has a wide dose range: Groups of 6 BALB/c mice were immunized i.m. 1 time on day 0 with 30 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 25 μg of empty plasmid or 25 μg of pIRES-mIL-12 as a comparator adjuvant or decreasing doses of pCTA1. Enough empty plasmid was included to keep the total DNA delivered in each vaccination to at least 70 μg. An additional group had the vaccine DNA delivered i.m. with electroporation. Day 14 splenocytes were stimulated with SIVmac239gag or HIVBaLgp120 peptide pools for IFN-γ ELISpot assays. The error bars represent the standard errors of the means. The P values (compared to the no adjuvant group) were calculated with a Students T Test using SigmaPlot v12 software. NS stands for not significantly different from the unadjuvanted control. The results shown are from a single experiment performed.
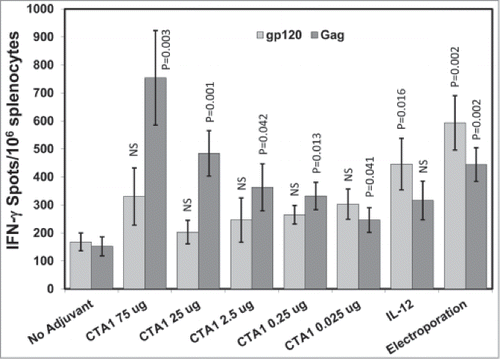
Figure 3. CTA1 has complementary adjuvant effects with IL-12 and GM-CSF: Groups of 6 BALB/c mice were immunized i.m. 1 time on day 0 with 30 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 40 μg of empty plasmid, 40 μg of pCTA1, 40 μg of pIL-12, 40 μg of pGM-CSF or 20 μg of pCTA1 + 20 μg of pIL-12 or 20 μg of pCTA1 + 20 μg of pGM-CSF. Day 14 splenocytes were stimulated with SIVmac239gag or HIVBalgp120 peptide pools for IFN-γ ELISpot assays. The error bars represent the standard errors of the means. The P values (compared to the no adjuvant group) were calculated with a Students T Test using SigmaPlot v12 software. NS stands for not significantly different from the unadjuvanted control. The results shown are from a single experiment of 3 performed.
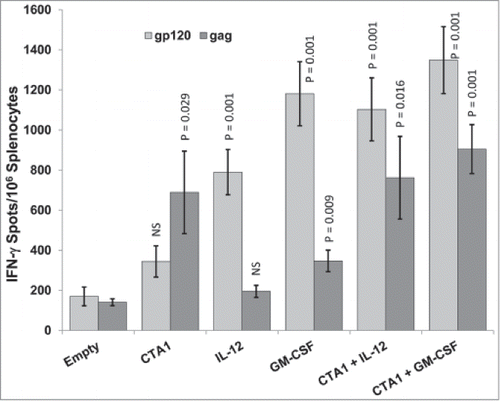
Figure 4. Complementary adjuvant effects of LTA1 with IL-12: Groups of 5 BALB/c mice were immunized i.m. on days 0 and 21 with 25 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 30 μg of empty plasmid, 30 μg of pLTA1, 30 μg of pIL-12 or 15 μg of pLTA1 + 15 μg of pIL-12. Day 35 splenocytes were stimulated with SIVmac239gag or HIV-BaLgp120 peptide pools for IFN-γ ELISpot assays and for supernatant cytokine analysis. (A) Splenocytes assayed by IFN-γ ELISpots. (B) Supernatants from peptide stimulated splenocytes plated at 8 × 105 cells/well analyzed by IFN-γ ELISA. (C) Supernatants from peptide stimulated splenocytes plated at 8 × 105 cells/well analyzed by Granzyme B ELISA. The error bars represent the standard errors of the means. The P values in panel (A) (compared to the no adjuvant group) were calculated with a Students T Test using SigmaPlot v12 software. The P values in panels (B and C)(compared to the no adjuvant group) were calculated with a Mann-Whitney Rank Sum-test using SigmaPlot v12 software. NS stands for not significantly different from the unadjuvanted control. The results shown are froma single experiment of 2 performed.
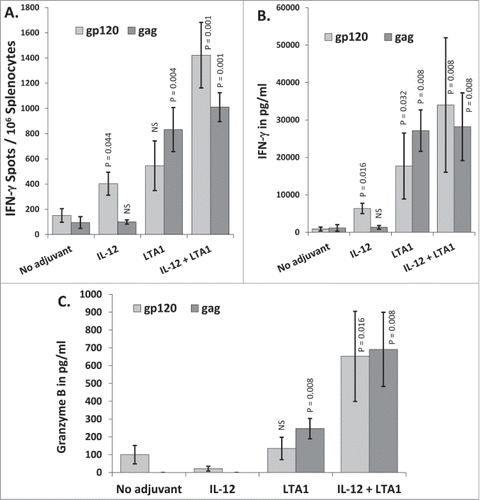
Figure 5. LTA1 evokes IL-17 secretion: Groups of 5 BALB/c mice were immunized i.m. on days 0 and 21 with 25 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 30 μg of empty plasmid, 30 μg of pLTA1 or 30 μg of pIL-12. Day 35 splenocytes were stimulated with SIVmac239gag or HIV-BaLgp120 peptide pools for IFN-γ ELISpot assays. Supernatants from peptide stimulated splenocytes plated a 1 × 105 cells/well were pooled within groups and analyzed by CBA. (A) Splenocytes assayed by IFN-γ ELISpots. (B) Supernatants from peptide stimulated splenocytes analyzed by multi-analyte CBA. C. Pooled supernatants from peptide stimulated splenocytes analyzed for IL-17 concentration by multi-analyte CBA. The error bars represent the standard errors of the means. The P values (compared to the no adjuvant group) were calculated with a Students T Test using SigmaPlot v12 software. NS stands for not significantly different from the unadjuvanted control. The results shown are from a single experiment of 2 performed.
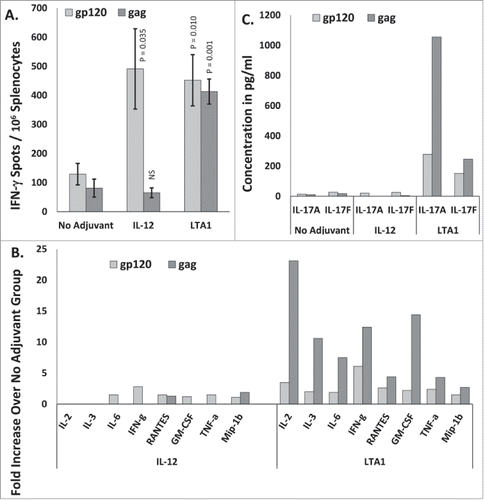
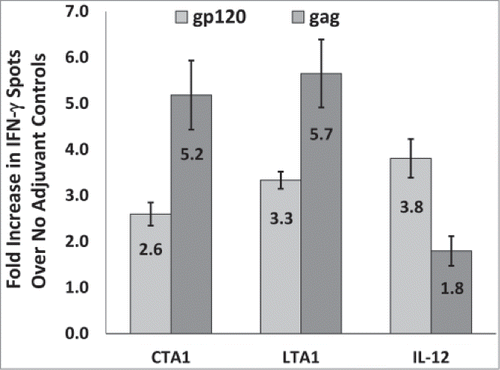
Figure 6. CTA1 and LTA1 preferentially enhance cellular responses to gag over gp120: The results from 9 independent mouse studies for CTA1, 6 independent studies for LTA1 and 9 independent studies for IL-12 were compared. Each study had groups of 5 BALB/c mice immunized i.m. on days 0 and 14 or days 0 and 21 with 25 μg of pSIV-gag and 15 μg of pHIV-gp120 plus either 25 μg of empty plasmid, pLTA1 or pIL-12. Day 28 or day 35 splenocytes were stimulated with SIVmac239gag or HIV-BaLgp120 peptide pools for IFN-γ ELISpot assays. For each study, the average gp120 and gag-specific IFN-γ ELISpot responses were determined for the no adjuvant groups and the CTA1, LTA1 or IL-12 adjuvanted groups. Within each study, the average fold increases for the gp120 (light gray bars) and gag (dark gray bars)-specific ELISpot responses were calculated by dividing the mean gp120 and gag-specific ELISpot responses from the adjuvanted groups by the corresponding mean gp120 and gag-specific ELISpot responses from the unadjuvanted groups. The error bars represent the standard errors of the means. The P values (provided in the text) were calculated with a Students T test using SigmaPlot v12 software.