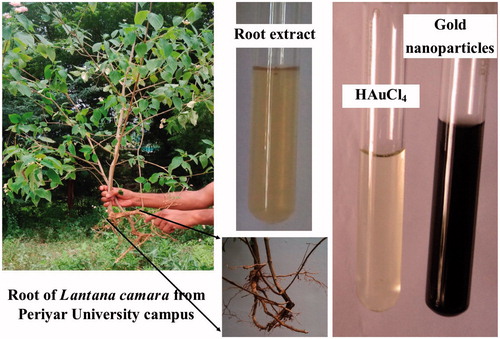
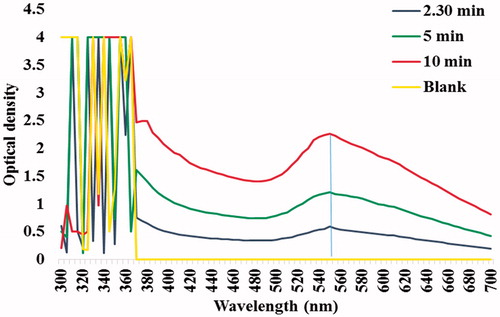
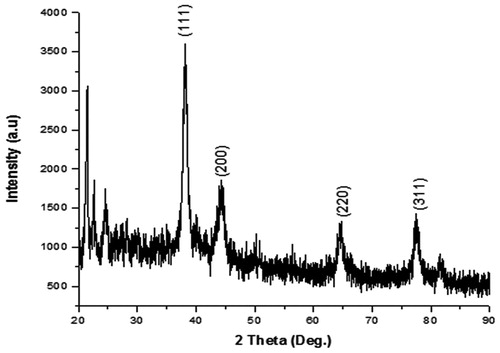
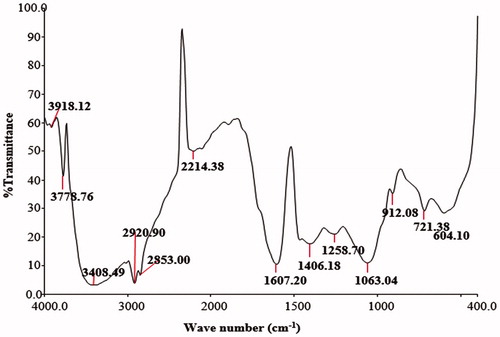
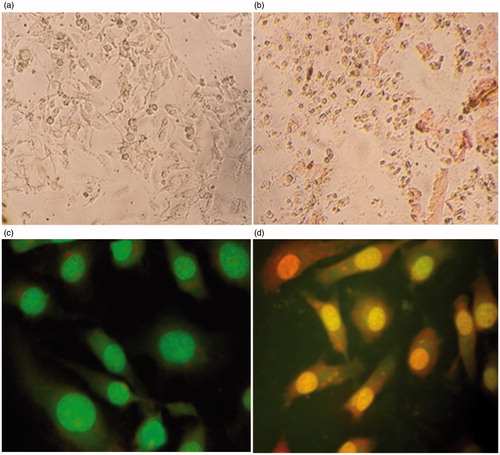
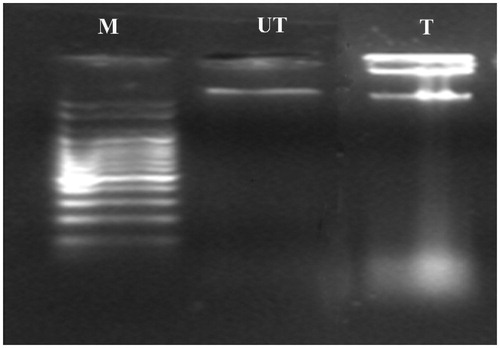
Figures & data
Figure 5. HR-TEM of Au NPs synthesized from root extract of L. camara. (a) and (b) the shapes and different sizes of Au NPs; (c) EDX analysis of Au NPs of L. camara; (d) SAED pattern showing that the nanoparticles are crystalline.
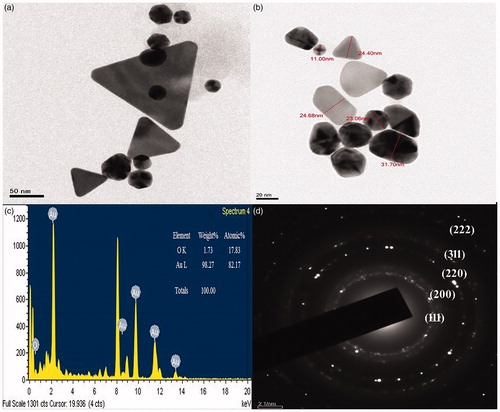
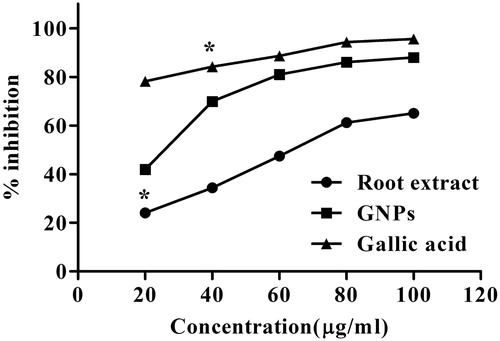
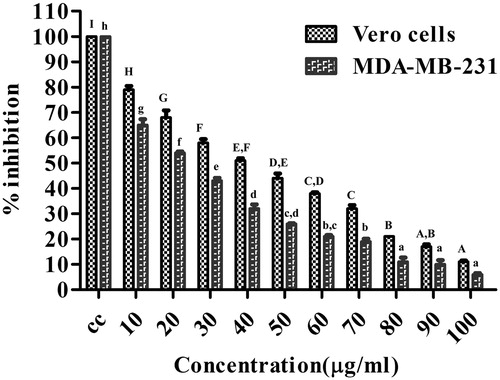
Figure 6. The DPPH scavenging assay of root extract, Au NPs, and Gallic acid.
DPPH scavenging ability of Au NPs vs. root extract and ascorbic acid. Values are mean of independent determinations. Two-way ANOVA, significant different from Au NPs: “*” p < 0.001.