Figures & data
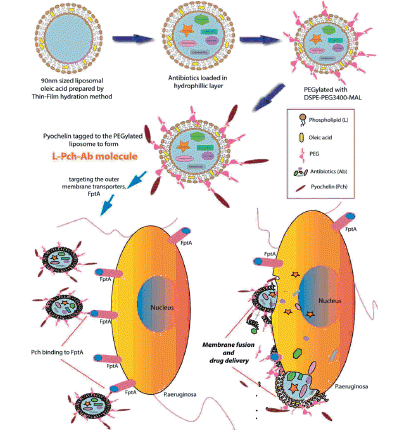
Scheme 2. PEGylated liposome complexes stabilized with pyochelin conjugates for specific targeting of P. aeruginosa.
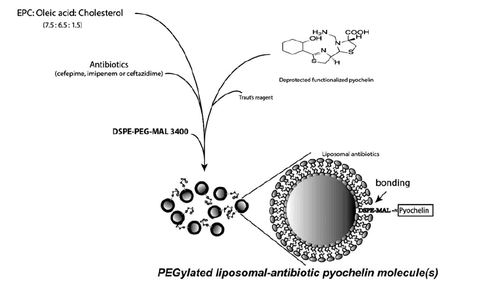
Table 1: Concentrations of F-Ab and L-Pch-Ab antibiotics (µg/ml) against ATCC 27853 and three clinical strains of P. aeruginosa (PS75, PS89 and PS103).
Figure 2. Characteristic TEM images of P. aeruginosa treated with various liposomal formulations at 4× MIC concentrations. (A) Bare liposomes, (B) MDRPa strain PS103, 4 h incubation with (C) L-CPM, (D) L-IPM, (E) L-CAZ, (F) L-Pch-CPM, (G) L-Pch-IPM, (H) L-Pch-CAZ and 6 h incubation with (I) L-Pch-CPM, (J) L-Pch-IPM and (K) L-Pch-CAZ.
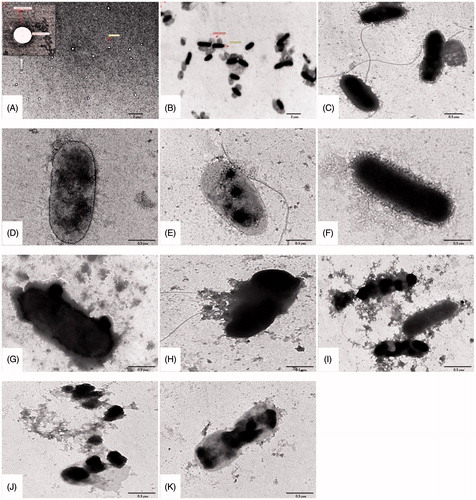
Figure 3. Time kill curves of (a–c) laboratory strain – ATCC 27853, (d–f) sensitive strain – PS75, (g–i) moderately resistant strain – PS89, (j–l) highly resistant strain – PS103.
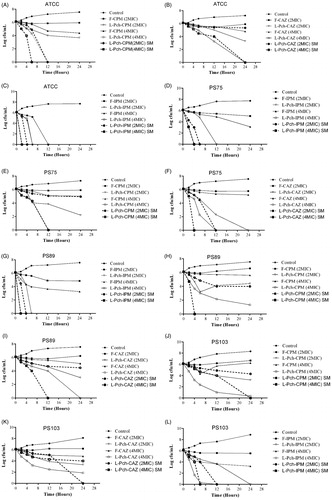
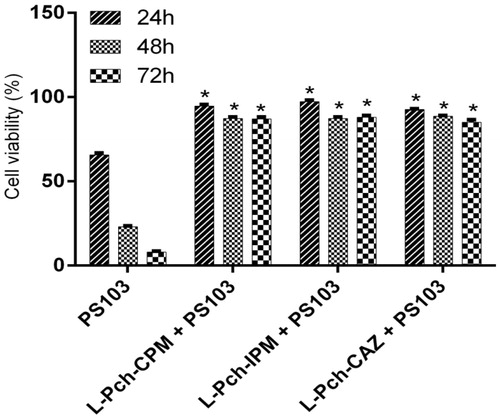
Figure 4. In vitro cytotoxicity of L-Pch-Ab formulations on HaCaT cells. HaCaT cells were treated with 4× MIC of L-Pch-CPM for 24, 48 and 72 h and cell viability was determined by MTT assay. Data expressed as a percentage of untreated control cells and are reported as the mean of three independent experiments ± SEM. *indicates a p value of <.05 compared to the corresponding resistant isolate PS103.