Figures & data
Table 1. Treatment groups.
Figure 1. Isolation and characterization of PDMSCs. (A) Fibroblastoid and uniform morphology of isolated PDMSCs after 1 month of culture. The typical morphology of adipocytes characterized by lipid droplet accumulation (oil red O) and formation of mineralized matrix revealed by alizarin red staining. Original magnification ×40. (B) Cell surface markers expression in flow cytometry of PDMSCs which are positive for CD105, CD90, CD44 and CD73 and negative for CD34 and CD45.
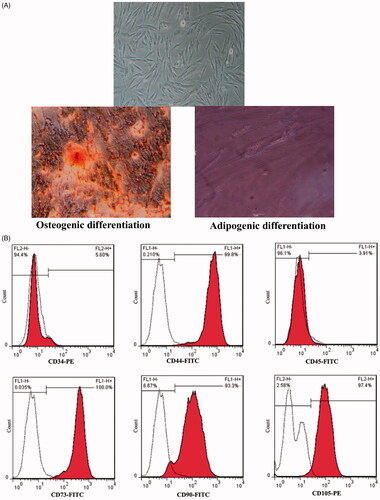
Figure 2. Fluorescence images of transduced PDMSCs and the expression of incorporated TRAIL gene. (A) The schematic image of the TRAIL construct gene. (B) Fluorescence images of GFP-PDMSCs and TRAIL-PDMSCs. (C) Western blot for the expressed TRAIL protein in the TRAIL-PDMSC. The original magnification is ×400.
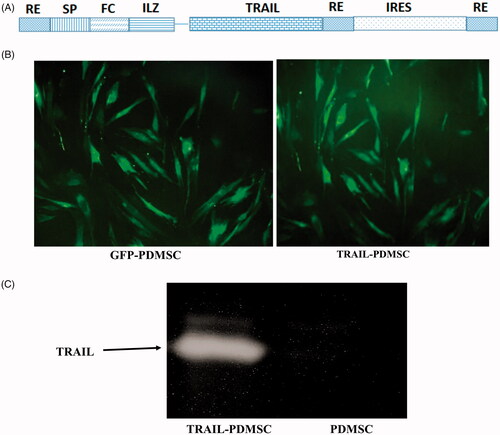
Figure 3. Curcumin-loaded chitosan nanoparticles characterization. (A) The average diameter of these nanoparticles is 652 ± 10 nm. (B) Different concentration of curcumin dissolved in ethanol was used to draw the standard curve. (C) The freeze-dried nanoparticles are stored at 4 °C. (D) The freeze-dried nanoparticles are easily dissolved in PBS and readily for injection.
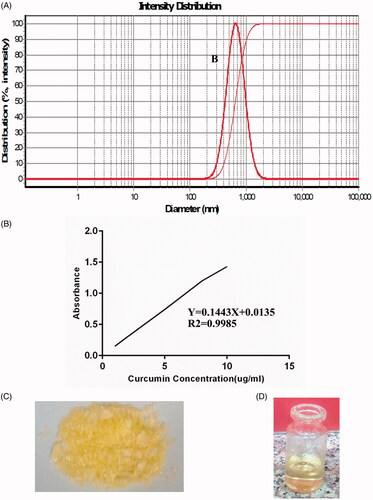
Figure 4. Tumour cells viability significantly reduced when CM of TRAIL-PMSCs is combined with Cu-NPs. (A) Trypan blue exclusion assay determined the cell viability of 4T1 cells in different treatment conditions after 24 h. Untreated cells were used as the control. (B) MTT assay results quantifying the viability of 4T1 cells under different treatment conditions and in three different time points. All results are expressed by mean ± SD. *p < .05, **p < .01, ***p < .001, ****p < .0001. CM: conditioning media; TMSCs: TRAIL-PDMSCs; GMSCs: GFP-PDMSCs.
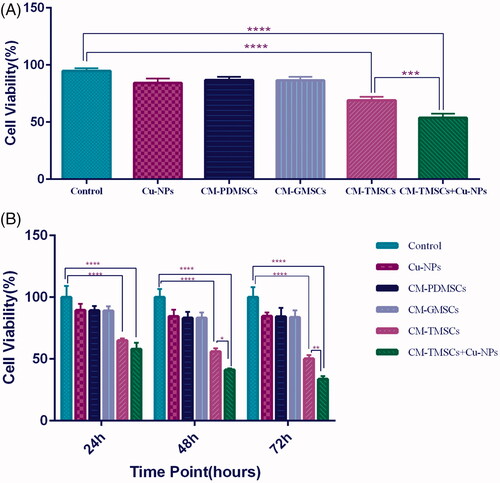
Figure 5. Anti-tumour effects of various treatment options in animal model of breast cancer. (A) 4T1 tumour cells were subcutaneously injected into right flank of BALB/c mice. (B) On the 28th day mice were sacrificed and tumour mass was removed. (C) H&E staining was performed on tumour tissues. (D) Tumour volume and weight were measured accurately as described in section “Materials and methods”. (E, F) TRAIL-PDMSCs when combined with Cu-NPs reduce tumour volume and weight to 0.44 and 0.34 as compared to control. All results are expressed by mean ± SD. *p < .05, ***p < .001, ****p < .0001. TMSC: TRAIL-PDMSCs; GMSC: GFP-PDMSCs.
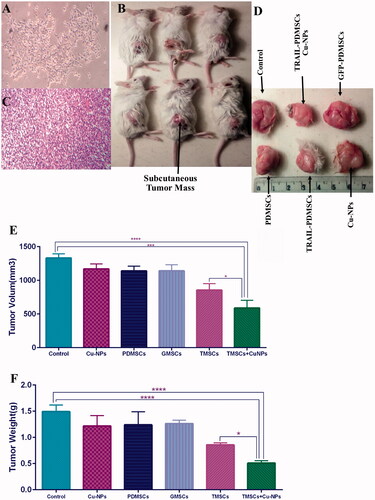
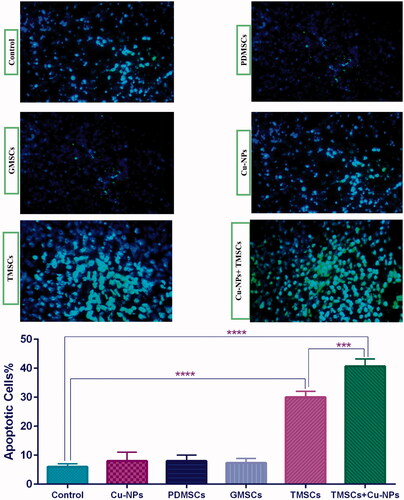
Figure 6. Analysis of apoptosis via TUNEL assay. (A) The TRAIL-PDMSCs + Cu-NPs group demonstrated a significant enhancement of TUNEL-positive nuclei. (B) The percentage of apoptotic cells between different experimental groups which shows no appreciable difference among the PDMSCs and GFP-PDMSCs groups. The number of apoptotic cells was counted in five random fields and avoiding areas of necrosis. Original magnification ×400. All results are expressed by mean ± SD.***p < .001, ****p < .0001. TMSC: TRAIL-PDMSCs; GMSC: GFP-PDMSCs.