Figures & data
Figure 1. A coevolutionary relationship represented using host and parasite phylogenies and a corresponding cophylogenetic reconstruction, which includes all four biological events.
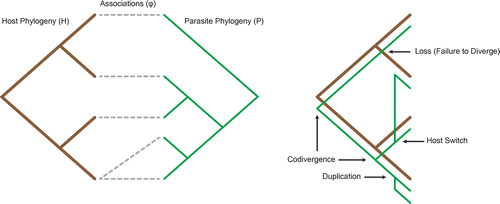
Figure 2. This is the algorithm that constructs a dynamic programming table for node mapping in . It calls a series of methods including three ‘Opt’ event functions (OptCodivergence, OptDuplication, OptHostSwitch) which query the preprocessed events. The fourth function is the AddMinCost function which ensures that only the minimum cost reconstruction for each host node is retained. As this function only compares two values at a time it runs in
.

Table 1. Comparing Jane 1 (node mapping), Jane 4 (edge mapping) and the newly proposed node mapping algorithm over six real data-sets using three different event cost vectors. The reported solutions are condensed into one column as all three algorithms reported the same map cost for all event cost vectors in all six test cases.
Figure 3. The running time of the original node mapping algorithm (Jane 1) and the new node mapping algorithm plotted on a logarithmic scale.
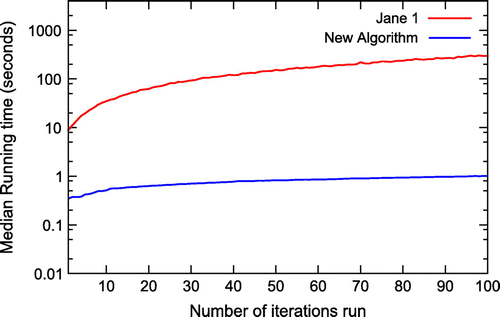
Table 2. The minimum, maximum and median running time for Jane 4 and the newly proposed node mapping algorithm over real data-set 6; host plants (Leguminosae) and phytophagous insects (Psylloidae); for 10,000 iterations repeated 1000 times.
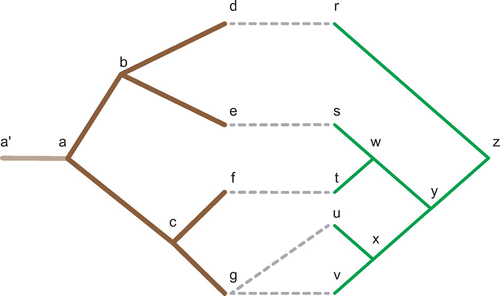
Figure 4. A coevolutionary relationship represented using the host and parasite phylogenies first shown in Figure . In this case, each node in the host and parasite tree is labelled to assist in the reconstruction described in Appendix 1. Of note node has been fixed to a timing interval before
which leads to the reconstruction described in Appendix 1 and an edge has been added to the root of
to map all parasite divergence events which occurred prior to this phylogeny.