Figures & data
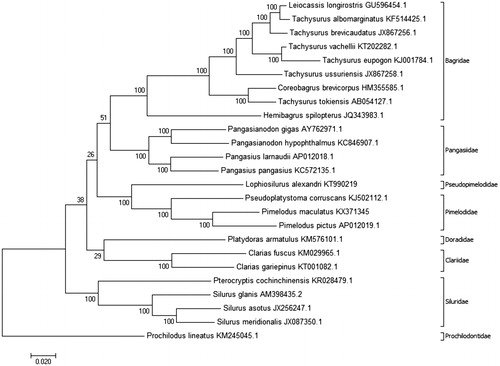
Figure 1. Molecular phylogenetic analysis inferred using the maximum likelihood method based on the Tamura–Nei model (Tamura & Nei Citation1993) with 1000 bootstrap replications. The analysis was carried out using complete mtDNA of Pimelodus maculatus (GenBank accession no. KX371345) and complete mtDNA of 25 other species: Leiocassis longirostris (GU596454.1), Tachysurus albomarginatus (KF514425.1), Tachysurus brevicaudatus (JX867256.1), Tachysurus vachellii (KT202282.1), Tachysurus eupogon (KJ001784.1), Tachysurus ussuriensis (JX867258.1), Coreobagrus brevicorpus (HM355585.1), Tachysurus tokiensis (AB054127.1), Hemibagrus spilopterus (JQ343983.1), Pangasianodon gigas (AY762971.1), Pangasianodon hypophthalmus (KC846907.1), Pangasius larnaudii (AP012018.1), Pangasius pangasius (KC572135.1), Lophiosilurus alexandri (KT990219), Pseudoplatystoma corruscans (KJ502112.1), Pimelodus pictus (AP012019.1), Platydoras armatulus (KM576101.1), Clarias fuscus (KM029965.1), Clarias gariepinus (KT001082.1), Pterocryptis cochinchinensis (KR028479.1), Silurus glanis (AM398435.2), Silurus asotus (JX256247.1), Silurus meridionalis (JX087350.1), and Prochilodus lineatus (KM245045.1). The phylogenetic tree with the highest log likelihood is shown, with the percentages of trees in which associated taxa clustered together shown next to the branches. The phylogenetic tree was rooted with P. lineatus (Characiformes, Prochilodontidae). The D-loop region was excluded from this analysis because it is considered to be highly variable (Gonder et al. Citation2007). The phylogenetic tree obtained was coherent with the phylogenetic studies of Sullivan et al. (Citation2006, Citation2013) and Lundberg et al. (Citation2011), which grouped species in their respective families; Pimelodidae and Pseudopimelodidae were grouped together in the superfamily Pimelodoidea. The analyses were conducted using MEGA7 software (Kumar et al. Citation2016).